A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Extraction of High Molecular Weight DNA from Microbial Mats
In This Article
Summary
We provide an improved protocol for extracting high molecular weight DNA from hypersaline microbial mats. Microbial cells are separated from the mat matrix prior to DNA extraction and purification. This enhances the concentrations, quality, and size of the DNA. The protocol may be used for other refractory samples.
Abstract
Successful and accurate analysis and interpretation of metagenomic data is dependent upon the efficient extraction of high-quality, high molecular weight (HMW) community DNA. However, environmental mat samples often pose difficulties to obtaining large concentrations of high-quality, HMW DNA. Hypersaline microbial mats contain high amounts of extracellular polymeric substances (EPS)1 and salts that may inhibit downstream applications of extracted DNA. Direct and harsh methods are often used in DNA extraction from refractory samples. These methods are typically used because the EPS in mats, an adhesive matrix, binds DNA2,3 during direct lysis. As a result of harsher extraction methods, DNA becomes fragmented into small sizes4,5,6.
The DNA thus becomes inappropriate for large-insert vector cloning. In order to circumvent these limitations, we report an improved methodology to extract HMW DNA of good quality and quantity from hypersaline microbial mats. We employed an indirect method involving the separation of microbial cells from the background mat matrix through blending and differential centrifugation. A combination of mechanical and chemical procedures was used to extract and purify DNA from the extracted microbial cells. Our protocol yields approximately 2 μg of HMW DNA (35-50 kb) per gram of mat sample, with an A260/280 ratio of 1.6. Furthermore, amplification of 16S rRNA genes7 suggests that the protocol is able to minimize or eliminate any inhibitory effects of contaminants. Our results provide an appropriate methodology for the extraction of HMW DNA from microbial mats for functional metagenomic studies and may be applicable to other environmental samples from which DNA extraction is challenging.
Protocol
1. Microbial Cell Extraction:
- Homogenize microbial mats with a sterile grinding pestle by mixing thoroughly. Place approximately all 30 g (wet weight) of homogenized mat material into sterile container of Waring blender, add about 100 mL of 1 M NaCl (or a concentration specific to sample in use), and blend three times at medium speed for 1 min with intermittent cooling in a -20°C freezer for 1 min. Transfer the slurry into a 250 mL centrifuge bottle and fill the remaining empty volume with 1 M NaCl. Prepare NaCl solution in autoclaved DI water and filter sterilize it.
- Further dislodge the microbial cells from the matrix by shaking (150 rpm) using a vortexer (Vortex-GenieR2) at room temperature for 30 min.
- Centrifuge at low speed (500x g) for 15 min at 4°C. Gently transfer the supernatant into a flask with minimum disturbance to the sediment.
- Using the pelleted sediment, repeat blending and steps 1.2 and 1.3 four additional times with the addition of fresh NaCl to each sediment pellet at the end of step 1.3. The supernatant from each NaCl extraction is transferred into a fresh flask.
- Combine the supernatants from the 5 cell extractions and centrifuge in aliquots of 200 mL at high speed (25,000x g) for 15 min at 4°C. Discard the supernatant, resuspend each cell pellet in 10 mL of 2% sodium hexametaphosphate, combine pellets and make up the volume to 200 mL with 2% sodium hexametaphosphate. Wash cells by shaking (150 rpm) at room temperature for 30 min. At the end of this step you should have only one tube of cells to proceed to the next step.
- Centrifuge at high speed (25,000x g) for 15 min at 4°C. Discard the supernatant, resuspend cell pellet in 200 mL of TE (50 mM EDTA), and wash cells by shaking (150 rpm) at room temperature for 10 min.
- Centrifuge at high speed (25,000x g) for 15 min at 4°C, discard the supernatant, resuspend in 15 mL TE (10 mM EDTA), and store 200-μL aliquots at -80°C until needed for DNA extractions.
2. DNA Extraction and Purification:
- Add 200 μL 5 M NaCl, 200 μL 10% SDS, and 100 μL 14.3 M β-mercaptoethanol to the 200-μL microbial cell aliquot. Mix gently by inverting 4-6 times.
- Subject the mixture to 3 rounds of freeze-thaw by submerging in liquid nitrogen for 2 min followed by thawing in a 65°C water bath for 5 min. Extend final thawing to 10 min.
- Add 200 μL 5 M potassium acetate (pH 5.5) and place on ice for 10 min. Centrifuge at 10,000x g for 10 min at 4°C and transfer the supernatant to a new tube using a wide-bore pipette tip. Add 3 μL of RNase A and incubate at 37°C for 1hr.
- Add equal volume of chloroform, shake briefly by inversion, and centrifuge at 15,000x g for 10 min at room temperature.
- Carefully transfer the top layer of supernatant into a new tube using a wide-bore pipette tip. Avoid pipetting the white interface layer located between the top and bottom layers. Repeat the chloroform-extraction procedure.
- Add an equal volume of chilled (at -20°C) isopropanol to the supernatant and incubate on ice for 30 min to precipitate DNA.
- Centrifuge at max speed (20,800x g) for 10 min at 4°C to pellet DNA.
- Discard the supernatant and wash DNA pellet with 1 mL of chilled (at -20°C) 70% ethanol.
- Centrifuge at max speed (20,800x g) for 10 min at 4°C, discard the supernatant, repeat 70% ethanol wash, and air-dry DNA pellet for 10 min. Repeat 70% ethanol wash.
- Resuspend DNA in 25 μL TE (10 mM EDTA), warm at 65°C for 5 min, combine into one tube if multiple tubes have been used, and make-up volume to 500 μL with TE.
- Add 500 μL ice-cold (or at 4°C) 20% polyethylene glycol (PEG) (prepared in 1.2 M NaCl). Mix gently by inversion, incubate on ice for 10 min, and centrifuge at maximum speed for 10 min at 4°C to pellet DNA. Remove and discard the supernatant and wash the pelleted DNA with 1 mL of chilled 70% ethanol, air-dry for 10 min, and resuspend in 30-50 μL TE or molecular grade water. Facilitate resuspension of DNA by warming at 65°C for 5 min and Store DNA at -80°C.
3. DNA Purity, Concentration and Size Determination:
- Determine DNA quality by measuring the A260/280 and A260/230 ratios using a Nanodrop 1000 spectrophotometer or any other appropriate instrumentation.
- Determine DNA concentration using Quant-iT dsDNA Assay kit according to the manufacturer’s instructions.
- Determine DNA size using pulse field gel electrophoresis. Prepare 1% agarose gel with 0.5X TBE and use the following parameters on the CHEF Mapper XA System; initial switch time of 0.35 s, a final switch time of 7.67 s, and 120° included angle for a gradient of 6.0 V/cm at a linear ramping factor. Stain gel with 1 x SYBR Green I for 30 min. Load between 700-1000 ng total DNA on gel. See Fig. 3 for a condensed form of the complete protocol.
Representative Results:
Cell extraction:
Sequential microbial cell extracts (supernatants) have shown that the turbidity of extracts decreases as the number of extractions increase. This suggests a reduction in the number of cells following each successive cell extraction. Important to note here is that as each additional cell extraction step provides the opportunity for contaminant introduction, the number of cell extractions should be minimized so that the final cell pellet is representative of the overall microbial community. Other sources of contamination were controlled by adopting adequate laboratory sterile techniques. For instance, the solutions were prepared in autoclaved DI water and filter sterilized. Containers were sterilized with alcohol, autoclaved, and treated under UV light and ultraviolet crosslinker.
DNA concentration and quality determination:
The protocol yielded approximately 2 μg of HMW DNA (35-50 kb) (Fig. 4) per gram of mat sample with an A260/280 ratio of 1.6, and an A260/230 ratio of 0.7 (Table 1). Although the A260/230 ratio appeared to be low, no inhibition of downstream molecular-based application such as PCR-amplification of the 16S rRNA genes was observed in a separate study7. It is important to note that DNA from hypersaline mats has two main sources of contamination; EPS and salts. It is therefore possible that traces of these contaminants may be influencing the A260/230 ratios despite the tremendous efforts to reduce their effects on down stream applications.
DNA size determination:
Pulse field gel electrophoresis employs a pulsating current flow with intermittent directional switches resulting in a DNA smear as shown in figure 3. Our protocol yielded a HMW DNA of approximately 35-50 kb. While some DNA size may be smaller than 30 kb, it is important to have part of the smear above 35 kb given that fosmid cloning requires ˜40 kb DNA inserts and larger DNA fragments provide greater access to intact biosynthetic pathways. In our studies, the DNA smear above 35 kb was excised and purified for large-insert vector cloning and other molecular applications.

Figure 1. Hypersaline microbial mat sampling site (Big Pond) located on Eleuthera, The Bahamas.

Figure 2. A cross-section of the hypersaline microbial mat used in this study. The microbial mat was obtained from a hypersaline pond located on Eleuthera, The Bahamas.
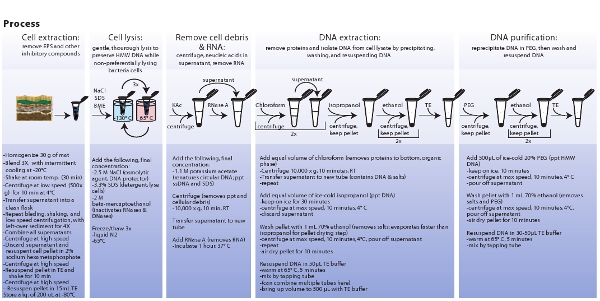
Figure 3. Schematic representation of procedures involved in microbial cell extraction, cell lysis, extraction, and purification of metagenomic DNA. Click here to view a larger image.
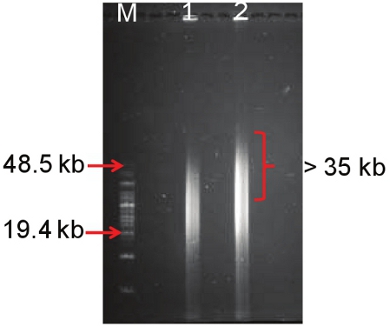
Figure 4. Molecular weight characterization of extracted DNA using pulse field gel electrophoresis. Lane M is a HMW marker, and lanes 1 and 2 are replicates of metagenomic DNA extracted from the Eleuthera hypersaline mat using the above protocol.
| Extraction Method | Concentration ng/g | A260/280 | A260/230 | |
| PEG | Rep 1 | 1806.1 | 1.64 | 0.76 |
| Rep 2 | 2010.8 | 1.61 | 0.75 |
Table 1. Measurement of concentration and quality of DNA extracted from microbial hypersaline mat.
Access restricted. Please log in or start a trial to view this content.
Discussion
Given that total cell removal from complex and highly diverse microbial mat samples is not practical, the primary concern is how well the extracted cells represent the overall microbial mat community. In a previous study, PCR-DGGE analysis of microbial 16S rRNA genes showed that the five cell removal steps used in this protocol extracts cells that are representative of the overall microbial mat community7. The actual number of cell extraction steps required to provide a cell pellet that is representative of th...
Access restricted. Please log in or start a trial to view this content.
Disclosures
No conflicts of interest declared.
Acknowledgements
This work was funded by the National Science Foundation Environmental Genomics Program (Grant No. EF-0723707).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| β-mercapt–thanol | Sigma-Aldrich | M3148 | |
| Polyethylene glycol 8000 | Promega Corp. | V3011 | 20% in 1.2 M NaCl |
| Potassium acetate | Fisher Scientific | Fisher Scientific | |
| Quant-iT dsDNA Assay kit | Invitrogen | Q33130 | |
| RNase | Epicentre Biotechnologies | MRNA092 | |
| Sodium Chloride | VWR | BDH8014 | Appropriate conc. |
| Sodium Dodecyl Sulfate | Fisher Scientific | 03-500-509 | 10% in water |
| sodium hexametaphosphate | EMD Millipore | SX0583-3 | 2% in water |
| TBE | Fisher Scientific | BP1333-1 | |
| CHEF Mapper XA System | Bio-Rad | 170-3670 | |
| NanoDrop 1000 spectrophotometer | Thermo Fisher Scientific, Inc. | ND-1000 | |
| Vortexer | Scientific Industries Inc. | ||
| Ultraviolet Crosslinker | UVP Inc. | ||
| Waring blender | Waring Laboratory | LB10S |
References
- Decho, A. W. Microbial biofilms in intertidal systems: an overview. Cont. Shelf Res. 20, 1257-1273 (2000).
- Dupraz, C., Visscher, P. T. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 13, 429-438 (2005).
- Steffan, R. J., Goksoyr, J., Boj, A. K., Atlas, R. M. Recovery of DNA from soils and sediments. Appl. Environ. Microbiol. 54, 2908-2915 (1988).
- Lee, Y. K., Kim, H. W., Liu, C. L., Lee, H. K. A simple method for DNA extraction from marine bacteria that produce extracellular materials. J. Microbiol. Methods. 52, 245-250 (2003).
- Roose-Amsaleg, C. L., Garnier-Sillam, E., Harry, M. Extraction and Purification of Microbial DNA from Soil and Sediment Samples. Appl. Soil Ecol. 18, 47-60 (2001).
- de Lipthay, J. R., Enzinger, C., Johnsen, K., Aamand, J., Sørensen, S. J. Impact of DNA extraction method on bacterial community composition measured by denaturing gradient gel electrophoresis. Soil Biol. Biochem. 36, 1607-1614 (2004).
- Bey, B. S., Fichot, E. B., Dayama, G., Decho, A. W., Norman, R. S. Extraction of High Molecular Weight DNA from Microbial Mats. Biotechniques. 49, 631-640 (2010).
- Kakirde, S. K., Parsley, L. C., Liles, M. R. Size does matter: Application-driven approaches for soil metagenomics. Soil Biology & Biochemistry. 42, 1911-1923 (2010).
- Rodon, M. R., August, P. R., Bettermann, A. D., Brady, S. F., Grossman, T. H., Liles, M. R., Loiacono, K. A., Lynch, B. A., MacNeil, I. A., Minor, C., Tiong, C. L., Gilman, M., Osburne, M. S., Clardy, J., Handelsman, J., Goodman, R. M. Cloning the Soil Metagenomic: A Strategy for Accessing the Genetic and Functional Diversity of Uncultured Microorganisms. Appl. Environ. Microbiol. 66, 2541-2547 (2000).
- Beja, O., Aravind, L., Koonin, E. V., Suzuki, M. T., Hadd, A., Nguyen, L. P., Jovanovich, S. B., Gates, C. M., Feldman, R. A., Spudich, J. L., Spudich, E. N., DeLong, E. F. Bacterial Rhodopsin: Evidence for a New Type of Phototrophy in the Sea. Science. 289, 1902-1906 (2000).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved