A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Pull-down of Calmodulin-binding Proteins
In This Article
Summary
Calmodulin (CaM) pull-down assay is an effective way to investigate the interaction of CaM with various proteins. This method uses CaM-sepharose beads for efficient and specific analysis of CaM-binding proteins. This provides an important tool to explore CaM signaling in cellular function.
Abstract
Calcium (Ca2+) is an ion vital in regulating cellular function through a variety of mechanisms. Much of Ca2+ signaling is mediated through the calcium-binding protein known as calmodulin (CaM)1,2. CaM is involved at multiple levels in almost all cellular processes, including apoptosis, metabolism, smooth muscle contraction, synaptic plasticity, nerve growth, inflammation and the immune response. A number of proteins help regulate these pathways through their interaction with CaM. Many of these interactions depend on the conformation of CaM, which is distinctly different when bound to Ca2+ (Ca2+-CaM) as opposed to its Ca2+-free state (ApoCaM)3.
While most target proteins bind Ca2+-CaM, certain proteins only bind to ApoCaM. Some bind CaM through their IQ-domain, including neuromodulin4, neurogranin (Ng)5, and certain myosins6. These proteins have been shown to play important roles in presynaptic function7, postsynaptic function8, and muscle contraction9, respectively. Their ability to bind and release CaM in the absence or presence of Ca2+ is pivotal in their function. In contrast, many proteins only bind Ca2+-CaM and require this binding for their activation. Examples include myosin light chain kinase10, Ca2+/CaM-dependent kinases (CaMKs)11 and phosphatases (e.g. calcineurin)12, and spectrin kinase13, which have a variety of direct and downstream effects14.
The effects of these proteins on cellular function are often dependent on their ability to bind to CaM in a Ca2+-dependent manner. For example, we tested the relevance of Ng-CaM binding in synaptic function and how different mutations affect this binding. We generated a GFP-tagged Ng construct with specific mutations in the IQ-domain that would change the ability of Ng to bind CaM in a Ca2+-dependent manner. The study of these different mutations gave us great insight into important processes involved in synaptic function8,15. However, in such studies, it is essential to demonstrate that the mutated proteins have the expected altered binding to CaM.
Here, we present a method for testing the ability of proteins to bind to CaM in the presence or absence of Ca2+, using CaMKII and Ng as examples. This method is a form of affinity chromatography referred to as a CaM pull-down assay. It uses CaM-Sepharose beads to test proteins that bind to CaM and the influence of Ca2+ on this binding. It is considerably more time efficient and requires less protein relative to column chromatography and other assays. Altogether, this provides a valuable tool to explore Ca2+/CaM signaling and proteins that interact with CaM.
Protocol
Refer to Figure 1 for a basic schematic of the procedure beginning with the homogenate. Estimated time from preparation of cellular extracts to elution of CaM-bound proteins is about six to seven hours.
1. Tissue preparation
- Inject organotypic hippocampal slices with a virus containing a plasmid expressing the recombinant protein of interest (in this example, green fluorescent protein (GFP)-tagged Ng) and allow tissue to express protein overnight.
- Approximately 12 to 18 hours after viral injection (depending on the viral expression time), prepare to collect the tissue. Add 1mL dissection buffer (10mM glucose, 4mM KCl, 26mM NaHCO3, 233mM sucrose, 5mM MgCl2, 1mM CaCl2, and 0.1% phenol-red, gassed with 5% CO2 95% O2 ) to a petri dish. Transfer cultured tissue/insert to petri dish and add 2mL dissection buffer to the insert to submerge the tissue.
- Collect organotypic hippocampal tissue (between 5 and 10 slices) by gently scraping the tissue free of the insert membrane using a scalpel. Specifically removing a particular region of interest (e.g. CA1) is also an option. Transfer the suspended tissue to a 1.5mL microcentrifuge tube using an inverted Pasteur pipette.
- Centrifuge samples at 1,500 rcf for 1 min to separate tissue from the dissection buffer. Carefully remove the supernatant by aspiration. Make sure not to disturb the pellet.
- For each slice used, add 30 to 60 μL of homogenization buffer (150mM NaCl, 20mM Tris pH 7.5, 1mM DTT, 1μg/mL leupeptin, 1μg/mL chemostatin, 1μg/mL antipain, 1μg/mL pepstatin, and 1% Triton X-100) to the tissue and homogenize thoroughly with pestle.
- In order to remove cellular debris, centrifuge the remaining homogenate at 1,100 rcf for 10 min and carefully remove the supernatant by aspiration while avoiding contamination from the pellet.
- Take 10% of the supernatant as a sample of the input (sample 1). Store the remaining supernatant on ice during preparation of the CaM-sepharose beads for use in step 3.
Note: The tissue used here are organotypic hippocampal slices. However, one could use dissociated neurons or any other cell culturing system. In such a case, begin at step 1.4 after collecting your tissue in the appropriate manner.
2. Preparation of beads for pull-down
In handling the beads, it is important to salvage the beads and maximize the efficiency of the reactions by preventing the beads from drying on the sides of the tube. To do so, it is best to rotate your tubes on their side, allowing solution to wet the beads on the walls of the tube, immediately before centrifugation.
- For each pull-down, pipette 400 μL of suspended Calmodulin-Sepharose beads into a 2 mL microcentrifuge tube with a relatively flat-bottom to maximize surface area and interaction of your solutions with the beads during your incubations.
- Centrifuge beads at 21,000 rcf for 30 seconds and carefully remove the supernatant by aspiration. Make sure not to disturb the beads.
- To wash the beads, add 100 μL of the respective homogenization buffer containing either 2 mM CaCl2 to those being used to pull-down Ca2+-CaM binding proteins and 2 mM EDTA (a known Ca2+ chelator) for beads pulling down ApoCaM binding proteins. Gently tap the tube to re-suspend beads and centrifuge at 1,500 rcf for 1 min. Carefully remove the supernatant by aspiration, making sure not to disturb the beads.
Note: For all the aspiration steps, it is recommended to use a pipette tip that has a fine opening (e.g. gel loading tips) to allow removal of solution without removing beads.
3. CaM-sepharose binding of proteins
- Split your supernatant into two conditions containing an equal volume. Depending on your condition, add the appropriate amount of CaCl2 or EDTA to your supernatant up to 2 mM concentration for each.
- Add supernatant from step 1.7 to beads washed in corresponding homogenization buffer. Gently tap the tube to mix.
- Incubate samples at 4°C for 3 hrs on a shaker. Re-suspend beads every 30 min or so to increase efficiency of binding.
- Centrifuge tube containing samples and beads at 1,500 rcf for 3 min.
- Take 50μL of supernatant as a sample of unbound protein (sample 2) and carefully remove the remaining supernatant by aspiration and discard.
- Wash beads three times as described in step 2.3 using 100μL of respective homogenization buffer.
4. Elution
- Add 50μL of elution buffer (50mM Tris-HCl pH 7.5, and 150mM NaCl) containing the opposite condition (10mM CaCl2 or 10mM EDTA) to the beads. For example, samples that were homogenized and bound in buffer containing Ca2+ are eluted in elution buffer containing EDTA and vice-versa.
Optional: Warming the elution buffer to 37°C before adding to beads may enhance the yield.
- Incubate solution with beads at room temperature for 30 min on a shaker. Mix the beads by gently tapping the tube about every 5 min.
- Centrifuge beads at 1,500 rcf for 3 min and carefully remove the 50μL of supernatant for the bound protein (sample 3) by aspiration. Make sure not to disturb the beads.
- Add protein loading buffer to all samples (i.e. homogenate, unbound and bound protein as well as those still bound to the beads).
Note: To maximize elution (especially in the case of inefficient elution), add 50μL of the corresponding elution buffer (e.g. adding EDTA containing buffer to beads bound in CaCl2) to the beads before heating samples to help elute any remaining bound protein and repeat steps described in 4.3 to remove remaining bound proteins.
5. SDS-PAGE and western blot
Conduct SDS-PAGE and analyze using western blot by probing for your protein of interest and probe for a protein known to bind CaM in the opposite condition as a positive control.
6. Representative Results
Figure 2B shows an example of a CaM-pull-down assay testing the CaM binding of GFP-tagged Ng compared to endogenous Ng. To do so, GFP-Ng was overexpressed in our organotypic hippocampal slices overnight and the tissue was homogenized. The homogenate was incubated with CaM-sepharose beads in the presence of either Ca2+ or EDTA. Homogenate input shows that the GFP-Ng was expressed in addition to endogenous Ng and Ca2+/CaM-dependent kinase II (CaMKII). As expected based on the known binding of endogenous Ng (illustrated in Fig. 2A), GFP-tagged Ng was eluted in the absence of Ca2+ (EDTA bound protein) and unbound in the presence of Ca2+ (Fig. 2B). In contrast, the control, CaMKII, was eluted only in the presence of Ca2+ (bound protein) and was unbound in its absence (EDTA). This shows that the CaM beads were functioning properly and the elutions were efficient. Most importantly, this shows that GFP-Ng binds to ApoCaM in a similar fashion to the endogenous form, suggesting that the GFP tag did not alter the function of our recombinant protein.
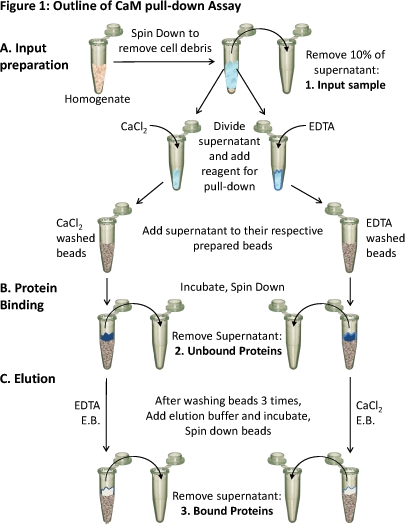
Figure 1. Outline of the CaM pull-down assay
(A) Tissue homogenate is spun down to remove cellular debris. About 10% of the supernatant is taken as a sample of the input (1). The remaining supernatant is divided equally for the different conditions and the appropriate reagents (CaCl2 or EDTA) are added to test binding in those conditions. Each supernatant (containing either CaCl2 or EDTA) is loaded onto the respectively prepared CaM-sepharose beads and (B) incubated to allow binding. Unbound proteins are removed (2) and (C) the bound proteins (3) are eluted off of the beads using elution buffer (E.B.) containing the opposite condition as the binding. The protein composition of these three protein samples can be analyzed using SDS-PAGE and western blot analysis.
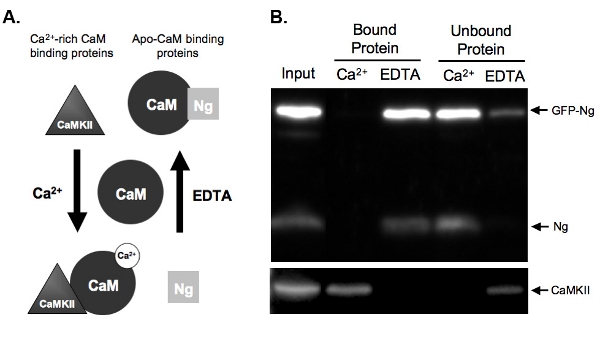
Figure 2. A.) Schematic of Ca2+-dependent CaM binding and elution in pull-down assay Examples are given of two types of protein that bind CaM in a Ca2+-dependent manner.
Neurogranin (Ng) represents proteins that bind apo-CaM and CaMKII represents proteins that bind to Ca2+-rich CaM. CaM is shown in its dissociated state prior to incubation with the homogenate proteins. Once incubated under conditions of high Ca2+ concentrations (2 mM) or in the presence of a Ca2+ chelator, EDTA (2 mM), the proteins will bind to CaM accordingly. Ng binds CaM in the EDTA condition as there is little to no Ca2+ present, and would be eluted off the CaM-sepharose beads in the presence of Ca2+. CaMKII, however, would bind to CaM in the presence of high amounts of Ca2+ and would dissociate once the Ca2+ was chelated.
B.) Results from example CaM pull-down assay. This figure demonstrates the expected end result of a CaM-sepharose pull down with the samples probed for Ng and CaMKII. Both the endogenous Ng and GFP-Ng are present in the lanes of proteins bound to CaM in the presence of EDTA. No Ng is bound when samples are incubated with CaM in the presence of Ca2+, demonstrating that Ng only binds apo-CaM. Our positive control, CaMKII, on the other hand, binds to CaM only in the presence of Ca2+.
Discussion
The provided protocol utilizes CaM-sepharose beads to investigate the Ca2+-dependence of CaM-binding proteins. Many proteins bind CaM in a Ca2+-dependent manner. These interactions are of great importance given the number of CaM-binding proteins and their critical role in many signaling pathways. In this protocol, CaM-sepharose beads are used to separate CaM-binding proteins from tissue homogenate in the presence or absence of Ca2+. The results of this simple approach will further the...
Disclosures
No conflicts of interest declared.
Acknowledgements
The authors would like to thank Tiffany Cherry in her help in optimizing this protocol. This work was funded by National Institute of Aging (AG032320) as well as Advancing a Healthier Wisconsin.
Materials
| Name | Company | Catalog Number | Comments |
| Calmodulin-Sepharose beads | GE Healthcare | 17-0529-01 | |
| Anti-CamKII alpha | Sigma-Aldrich | C6974 | |
| Anti-neurogranin | EMD Millipore | 07-425 | |
| Gel Loading Pipet Tips | Fisher Scientific | 02-707-138 | Use for aspiration of supernatants |
| Microcentrifuge tubes (2.0 mL) | Fisher Scientific | 05-408-146 | Use for all steps involving calmodulin-sepharose beads |
References
- Vincenzi, F. F. Calmodulin in the regulation of intracellular calcium. Proc. West Pharmacol Soc. 22, 289-294 (1979).
- Cheung, W. Y. Calmodulin plays a pivotal role in cellular regulation. Science. 207, 19-27 (1980).
- Zhang, M., Tanaka, T., Ikura, M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 2, 758-767 (1995).
- Alexander, K. A., Wakim, B. T., Doyle, G. S., Walsh, K. A., Storm, D. R. Identification and characterization of the calmodulin-binding domain of neuromodulin, a neurospecific calmodulin-binding protein. J. Biol. Chem. 263, 7544-7549 (1988).
- Huang, K. P., Huang, F. L., Chen, H. C. Characterization of a 7.5-kDa protein kinase C substrate (RC3 protein, neurogranin) from rat brain. Arch. Biochem. Biophys. 305, 570-580 (1993).
- Bahler, M., Rhoads, A. Calmodulin signaling via the IQ motif. FEBS Lett. 513, 107-113 (2002).
- Routtenberg, A. Protein kinase C activation leading to protein F1 phosphorylation may regulate synaptic plasticity by presynaptic terminal growth. Behav. Neural. Biol. 44, 186-200 (1985).
- Zhong, L., Cherry, T., Bies, C. E., Florence, M. A., Gerges, N. Z. Neurogranin enhances synaptic strength through its interaction with calmodulin. EMBO J. 28, 3027-3039 (2009).
- Needham, D. M. Myosin and adenosinetriphosphate in relation to muscle contraction. Biochim. Biophys. Acta. 4, 42-49 (1950).
- Hathaway, D. R., Adelstein, R. S. Human platelet myosin light chain kinase requires the calcium-binding protein calmodulin for activity. Proc. Natl. Acad. Sci. U.S.A. 76, 1653-1657 (1979).
- Fukunaga, K., Yamamoto, H., Matsui, K., Higashi, K., Miyamoto, E. Purification and characterization of a Ca2+- and calmodulin-dependent protein kinase from rat brain. J. Neurochem. 39, 1607-1617 (1982).
- Yang, S. D., Tallant, E. A., Cheung, W. Y. Calcineurin is a calmodulin-dependent protein phosphatase. Biochem. Biophys. Res. Commun. 106, 1419-1425 (1982).
- Huestis, W. H., Nelson, M. J., Ferrell, J. E. J. Calmodulin-dependent spectrin kinase activity in human erythrocytes. Prog. Clin. Biol. Res. 56, 137-155 (1981).
- Yamniuk, A. P., Vogel, H. J. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27, 33-57 (2004).
- Zhong, L., Kaleka, K. S., Gerges, N. Z. Neurogranin phosphorylation fine-tunes long-term potentiation. Eur. J. Neurosci. 33, 244-250 (2011).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved