A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Isolation of Ribosome Bound Nascent Polypeptides in vitro to Identify Translational Pause Sites Along mRNA
In This Article
Summary
A technique to identify translational pause sites on mRNA is described. This procedure is based on isolation of nascent polypeptides accumulating on ribosomes during in vitro translation of a target mRNA, followed by the size analysis of the nascent chains using a denaturing gel electrophoresis.
Abstract
The rate of translational elongation is non-uniform. mRNA secondary structure, codon usage and mRNA associated proteins may alter ribosome movement on the messagefor review see 1. However, it's now widely accepted that synonymous codon usage is the primary cause of non-uniform translational elongation rates1. Synonymous codons are not used with identical frequency. A bias exists in the use of synonymous codons with some codons used more frequently than others2. Codon bias is organism as well as tissue specific2,3. Moreover, frequency of codon usage is directly proportional to the concentrations of cognate tRNAs4. Thus, a frequently used codon will have higher multitude of corresponding tRNAs, which further implies that a frequent codon will be translated faster than an infrequent one. Thus, regions on mRNA enriched in rare codons (potential pause sites) will as a rule slow down ribosome movement on the message and cause accumulation of nascent peptides of the respective sizes5-8. These pause sites can have functional impact on the protein expression, mRNA stability and protein foldingfor review see 9. Indeed, it was shown that alleviation of such pause sites can alter ribosome movement on mRNA and subsequently may affect the efficiency of co-translational (in vivo) protein folding1,7,10,11. To understand the process of protein folding in vivo, in the cell, that is ultimately coupled to the process of protein synthesis it is essential to gain comprehensive insights into the impact of codon usage/tRNA content on the movement of ribosomes along mRNA during translational elongation.
Here we describe a simple technique that can be used to locate major translation pause sites for a given mRNA translated in various cell-free systems6-8. This procedure is based on isolation of nascent polypeptides accumulating on ribosomes during in vitro translation of a target mRNA. The rationale is that at low-frequency codons, the increase in the residence time of the ribosomes results in increased amounts of nascent peptides of the corresponding sizes. In vitro transcribed mRNA is used for in vitro translational reactions in the presence of radioactively labeled amino acids to allow the detection of the nascent chains. In order to isolate ribosome bound nascent polypeptide complexes the translation reaction is layered on top of 30% glycerol solution followed by centrifugation. Nascent polypeptides in polysomal pellet are further treated with ribonuclease A and resolved by SDS PAGE. This technique can be potentially used for any protein and allows analysis of ribosome movement along mRNA and the detection of the major pause sites. Additionally, this protocol can be adapted to study factors and conditions that can alter ribosome movement and thus potentially can also alter the function/conformation of the protein.
Protocol
1. DNA Template Preparation and in vitro Transcription
- The gene of interest is cloned under T7 and/or e.g. SP6 transcriptional promoter.
- For in vitro transcription the template DNA is linearized with an appropriate restriction enzyme cutting downstream of the ORF stop codon and/or mRNA 3' end. One needs to verify complete linearization of the plasmid DNA by running the restriction digestion product on agarose gel electrophoresis.
- The linearized plasmid is used for in vitro transcription reaction. Different concentration of template DNA can be tested to identify optimum DNA concentration required for in vitro transcription. Generally, with Ambion's MEGAscript High yield Transcription Kit (Ambion/Life Technologies, Grand Island, NY), 1 μg linearized DNA yields 40-60 μg of mRNA. In vitro transcription is done following manufacturer's instruction (Ambion/Life Technologies, Grand Island, NY).
- Following in vitro transcription the mRNA is purified by lithium chloride precipitation according to manufacturer's instruction (Ambion's MEGAscript High yield Transcription Kit).
- mRNA integrity is further verified by electrophoresis on acrylamide or agarose gels.
2. In vitro Cell Free Translation
For in vitro translation using Rabbit Reticulocyte Lysate, RRL (Promega, Madison, WI) follow the steps below:
- Prepare 100 μl in vitro translation reaction following manufacturer's instruction. Briefly, in nuclease free water add 2 μl amino acid mixture minus methionine (1 mM), 10 units of RNase Inhibitor (Invitrogen/Life Technologies, Grand Island, NY), 20 μCi of radioactive [35S]-Methionine (MP Biomedicals, Solon, OH), 70 μl of Rabbit Reticulocyte Lysate (Promega, Madison, WI).
- Incubate the reaction at 30 °C for 5 min to pre warm the translation reaction. Add 2 μg of mRNA to the translation reaction and incubate at 30 °C for 5 min.
- Stop the reaction by placing the translation reaction on ice.
3. Isolation of Nascent Polypeptides from in vitro Translation Reaction
- To isolate nascent polypeptide bound to ribosomes (polysome), translation reaction is layered on top of 4.5 ml of 30% glycerol in 10 mM Tris-HCl buffer pH 7.6, containing 100 mM KCl, 10 mM MgCI2, and centrifuged at 100,000 X g in a TLA-110 rotor (Beckman Coulter, Inc, Brea, CA) for 1 hr at 4 °C.
- The supernatant is further carefully removed.
- The polysomal pellet containing nascent polypeptides is resuspended in a small volume (10-15 μl) of 1 mM Tris-HCl buffer pH 7.6, containing 0.5 mg/ml ribonuclease A (Invitrogen/Life Technologies, Grand Island, NY), and incubated for 30 min at 37 °C. In order to enhance the hydrolysis of the peptidyl-tRNA ester bond, NaOH is added to a final concentration of 10 mM, and the incubation is continued for additional 30 min.
In vitro translation reaction assembled without mRNA and performed under the same conditions (followed by isolation of nascent peptides as has been described) would serve as a major control. Treatment of the translation reaction(s) with puromycin before subjecting the extract to density gradient centrifugation may serve as an additional control.
4. Resolving the Nascent Polypeptide on Tris-tricine SDS PAGE
- The isolated nascent polypeptides are resolved and analyzed on Tris- tricine SDS-PAGE. Please note that the amount of the material loaded on the gel will dependent on the efficiency of protein labeling, efficiency on protein translation and many others parameters. The amount of loaded material should be adjusted empirically to allow the best visualization and separation of individual nascent polypeptide chains.
- After electrophoresis gels are fixed, dried using vacuum gel dyer and subjected to autoradiography. The distribution of nascent polypeptides are observed using phosphoimaging.
5. Representative Results
Bovine Gamma-B Crystallin was translated in Rabbit Reticulocyte Lysate as well as in E. coli S30 Extract Systems (Promega, Madison, WI) followed by isolation of nascent polypeptide chains. Figure 1 depicts steps involved in isolation of ribosome bound nascent chains. In the present study the aim was to test, if changing the environment of translation i.e. repertoire of tRNAs (known to be substantially different in mammalian (RRL) and prokaryotic (E. coli) systems due to differences in codon bias between the organisms) can alter ribosome movement along mRNA and effect the distribution of translation pause sites. Altered ribosome movement should lead to changes in the distribution of sizes of nascent polypeptides accumulating during translation, as well as their relative intensities. Relative intensities of the bands reflect the duration of the pause. The nascent polypeptides were isolated and resolved on 16.5%T , 6%C Tris-tricine SDS PAGE (Figure 2). Gamma-B Crystallin is a 20 kDa protein. The observation of nascent polypeptides of non-identical lengths and intensities in RRL and E. coli systems indicates that the movement of ribosomes along mRNA in these two systems follows different translation kinetics. For example, we observed a prominent band of 17.7 kDa in E. coli lysate and not in RRL system, suggesting that there is an additional translational pause site in the 3'- end region of mRNA (encoding C-terminal region of Gamma B Crystallin), when it is translated in E. coli system. We note that Gamma-B Crystallin harbors tandem Arg codons (AGG153 and AGA154) that are frequent in mammalian systems, but are known to be extremely rare and slowly translated in E. coli. A novel nascent peptide accumulating in E. coli system (~17.7 kDa Figure 2.2) is likely caused by ribosome pausing at these tandem rare codons. Please note that no bands can be observed in the absence of mRNA (Figure 2.3) and that puromycin treatment removes most of the nascent chains from polysomes (Figure 2.3). Prolonged treatment with puromycin (> 15 min) removes all the nascent chains (data not shown). This confirms that the observed polypeptide products are ribosome associated nascent chains, rather than mRNPs cosedimenting with polysomes.
As mentioned above, codon bias is organism as well as tissue specific. The representative example described here, clearly indicates that changing the environment of translation i.e. repertoire of tRNAs/codon bias can lead to altered movement of ribosome along the mRNA. Evidently, this simple technique can not only allow identification of the translational pause sites along mRNA, but also permits rapid comparison of the translation pause sites between different systems.
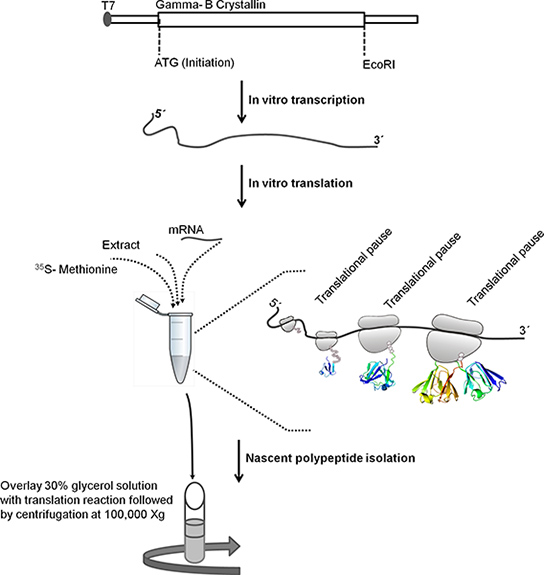
Figure 1. Schematic explaining the steps involved in isolating ribosome bound nascent polypeptides. Bovine Gamma-B Crystallin ORF sequence (528 base pairs) was cloned downstream of T7 promoter in pBSKS+. For in vitro transcription the template DNA was linearized by treating it with EcoRI. Linearized template was used for in vitro transcription. T7 promoter in DNA template is recognized by T7 RNA polymerase that transcribes the downstream Gamma-B Crystallin gene. mRNA is purified using lithium chloride precipitation method. The purified mRNA is used for in vitro translation. Following incubation, translation reaction is layered on top of 30% glycerol solution and centrifuged for 1 hr at 4 °C at 100,000 X g to isolate ribosome bound nascent polypeptides.
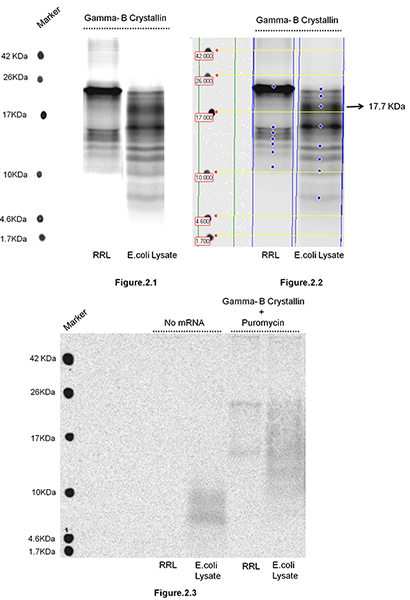
Figure 2. Isolation of Bovine Gamma-B Crystallin nascent polypeptides translated in Rabbit Reticulocyte Lysate (RRL) and E. coli Lysate. 2 μg mRNA was mixed in 100 μl reaction containing RRL or E. coli S30 Extract System (Promega, Madison, WI) according to manufacturer's instruction with 20 μCi [35S]-Methionine and incubated at 30 °C for 5 min. Following incubation the translation reaction was layered on top of 4.5 ml of 30% glycerol in 10 mM Tris-HCl buffer pH 7.6, containing 100 mM KCl, 10 mM MgCI2, and centrifuged for 1 hr at 4 °C and 100,000 X g in a TLA-110 rotor (Beckman Coulter, Inc, Brea, CA). The isolated polyribosome pellet was resuspended in 20 μl of 1 mM Tris-HCl pH 7.6, containing 0.5 mg/ml ribonuclease A (Invitrogen/Life Technologies, Grand Island, NY) followed by treatment with NaOH (as described in the protocol section). Nascent polypeptides were resolved on 16.5% T, 6% C Tris-Tricine SDS PAGE gel. The gel was dried and exposed for autoradiography. Following exposure the gel was scanned using GE Healthcare's Typhoon Imaging Scanner. Figure 2.1 shows the gel with nascent polypeptides isolated and resolved after translation reactions done in two different systems (RRL and E. coli). Figure 2.2 The gel was analyzed by Image Quant TL, v2005 software and used here as an example to show that the molecular weight and intensity of the nascent polypeptides accumulating in two systems can be easily analyzed and compared. Figure 2.3 shows two sets of controls: nascent polypeptides isolated and resolved on SDS PAGE after translation reactions were assembled without mRNA and/or after the translation reactions were treated with puromycin (5 min, final conc. 1 mM) before the reaction was subjected to centrifugation.
Discussion
For reproducible results, quality and concentration of the components used for in vitro transcription and translation reactions are critical. In the current study we have used commercially available kits and extracts that provide highly reproducible data, if handled carefully. However, translation-competent extracts can be prepared from the cell of one's choice, if needed. Quality of mRNA can affect the translation, so it is of utmost importance to test the integrity of mRNAs before using it during in vitro<...
Disclosures
No conflicts of interest declared.
Acknowledgements
This work was funded by Human Frontier Science Program grant RGP0024.
Materials
| Name | Company | Catalog Number | Comments |
| MEGAscript T7 High yield Transcription Kit | Ambion | AM1333 | |
| Ribonuclease Inhibitor | Invitrogen | 15518012 | |
| Trans [35S]-Label | MP Biomedicals | 0151006 | |
| Ribonuclease-A | Invitrogen | 12091 | |
| Rabbit Reticulocyte Lysate System, Nuclease Treated | Promega | L4960 | |
| E. coli S30 Extract System for Linear Templates | Promega | L1030 | |
| Centrifugation | Beckman Coulter | Optima TLX Ultracentrifuge | |
| Storage phosphor autoradiography | GE Healthcare | Typhoon 9410 variable mode imager | |
| Software for nascent polypeptide analysis | GE Healthcare | Image Quant TL, v2005 |
References
- Komar, A. A. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 34, 16-24 (2009).
- Sharp, P. M., Cowe, E., Higgins, D. G., Shields, D. C. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 16, 8207-8210 (1988).
- Dittmar, K. A., Goodenbour, J. M., Pan, T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221 (2006).
- Ikemura, T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2, 13-34 (1985).
- Wolin, S. L., Walter, P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 7, 3559-3569 (1998).
- Krasheninnikov, I. A., Komar, A. A., Adzhubei, I. A. Nonuniform size distribution of nascent globin peptides, evidence for pause localization sites, and a cotranslational protein-folding model. J. Protein Chem. 10, 445-454 (1991).
- Komar, A. A., Lesnik, T., Reiss, C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 462, 387-391 (1999).
- Komar, A. A., Jaenicke, R. Kinetics of translation of γ B crystallin and its circularly permutated variant in an in vitro cell-free system: possible relations to codon distribution and protein folding. FEBS Lett. 376, 195-198 (1995).
- Jha, S., Komar, A. A. Birth, life and death of nascent polypeptide chains. Biotechnol. J. 6, 623-640 (2011).
- Thanaraj, T. A., Argos, P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 5, 1594-1612 (1996).
- Kimchi-Sarfaty, C., Oh, J. M., Kim, I. W., Sauna, Z. E. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 315, 525-528 (2007).
- Schägger, H., von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368-379 (1987).
- Shirole, N., Balasubramanian, S., Yanofsky, C., Cruz-Vera, L. Isolation of Translating Ribosomes Containing Peptidyl-tRNAs for Functional and Structural Analyses. J. Vis. Exp. (48), e2498 (2011).
- Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S., Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324, 218-223 (2009).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved