A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Single Plane Illumination Module and Micro-capillary Approach for a Wide-field Microscope
In This Article
Summary
A module for single plane illumination microscopy (SPIM) is described which is easily adapted to an inverted wide-field microscope and optimized for 3-dimensional cell cultures. The sample is located within a rectangular capillary, and via a microfluidic system fluorescent dyes, pharmaceutical agents or drugs can be applied in small quantities.
Abstract
A module for light sheet or single plane illumination microscopy (SPIM) is described which is easily adapted to an inverted wide-field microscope and optimized for 3-dimensional cell cultures, e.g., multi-cellular tumor spheroids (MCTS). The SPIM excitation module shapes and deflects the light such that the sample is illuminated by a light sheet perpendicular to the detection path of the microscope. The system is characterized by use of a rectangular capillary for holding (and in an advanced version also by a micro-capillary approach for rotating) the samples, by synchronous adjustment of the illuminating light sheet and the objective lens used for fluorescence detection as well as by adaptation of a microfluidic system for application of fluorescent dyes, pharmaceutical agents or drugs in small quantities. A protocol for working with this system is given, and some technical details are reported. Representative results include (1) measurements of the uptake of a cytostatic drug (doxorubicin) and its partial conversion to a degradation product, (2) redox measurements by use of a genetically encoded glutathione sensor upon addition of an oxidizing agent, and (3) initiation and labeling of cell necrosis upon inhibition of the mitochondrial respiratory chain. Differences and advantages of the present SPIM module in comparison with existing systems are discussed.
Introduction
In addition to well established methods (confocal or multi-photon laser scanning microscopy1-4, structured illumination microscopy5,6) light sheet or single plane illumination microscopy (SPIM) has proven to be a valuable method of 3D imaging7,8,9. Of particular interest is its application to 3-dimensional cell cultures, e.g., multi-cellular tumor spheroids (MCTS), which are used increasingly for drug discovery research10,11. Furthermore, SPIM is a preferential method when even upon long-term exposure or repetitive measurements low light doses are required to maintain the sample’s viability, since for measurement of each plane of the sample only this plane is exposed to light. This is in contrast to other microscopy techniques, where for detection of each focal plane the whole sample is illuminated, so that upon recording of numerous planes the light dose sums up and may damage the sample12.
Light sheet microscopy or SPIM is based on illumination of the sample in perpendicular direction to the observation path either by use of a cylindrical lens or by scanning the exciting laser beam (for a review see Ref. 8). This often requires special sample chambers13,14 or matrices, e.g., agarose7,15, implemented in special high-cost microscopes. As an alternative to those systems a comparably simple illumination device for SPIM has been developed and adapted to a conventional inverted microscope16 (see Figure 1). It consists of a laser beam expanded to a diameter of 8 mm and focused by a cylindrical lens (focal length: 50 mm, numerical aperture: 0.08) to a light sheet of 6–10 µm thickness over a depth of field of about 100 µm. Samples are located in a rectangular capillary of 600–900 µm inner diameter placed in front of the microscope objective lens for fluorescence detection. These main features are presently completed and optimized by the use of advanced micro-capillary approaches for holding and for rotating the samples, the synchronous adjustment of the illuminating light sheet (in axial direction) and the objective lens used for fluorescence detection (identical optical path lengths of displacement require a correction of the mechanical feed), and the adaptation of a microfluidic system for application of fluorescent dyes, pharmaceutical agents or drugs, thus minimizing the required quantities and expenses.
Protocol
1. Cell Spheroid Growing and Incubation
- Experiment 1: Cell spheroids incubated with a chemotherapeutic drug
- Prepare agarose 1.5% in culture medium by adding 0.45 g of agarose to 30 ml culture medium (sufficient for 6 plates).
- Heat the mixture from step 1.1.1 to at least 80 °C while stirring it from time to time.
- Fill 50 µl of the heated mixture from step 1.1.2 into each well of a 96-well plate and let it solidify within 1–2 hr. Store the plates closed and covered in the refrigerator at 5 °C until use.
- Seed about 150 MCF-7 breast cancer cells in each well of the prepared 96-well plate in Dulbecco's Modified Eagle's Medium (DMEM) with Ham’s F-12 culture medium supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin.
- Grow spheroids for 5–7 days up to a diameter of about 200–300 µm in an incubator at 5% CO2 and 37 °C.
- Incubate the cell spheroids with the anthracycline antibiotic doxorubicin hydrochloride for 6 hr at a concentration ranging between 2 µM and 8 µM (in culture medium) in an incubator at 5% CO2 and 37 °C.
- Wash the cell spheroids with culture medium or Earle's Balanced Salt Solution (EBSS) prior to microscopy.
- Experiment 2: Oxidation process in spheroids expressing a redox sensor
- Seed about 400 U251-MG-L106 glioblastoma cells permanently transfected with the glutathione sensitive green fluorescent redox sensor Grx1-roGFP2 in agarose gel–coated wells (procedure described in steps 1.1.1–1.1.3) of a 96-well plate in DMEM culture medium supplemented with 10% FCS, 1% penicillin/streptomycin and hygromycin B (150 µg/ml).
- Grow spheroids for 5–7 days up to a diameter of about 200–300 µm in an incubator at 5% CO2 and 37 °C.
- Add 10 µl of hydrogen peroxide solution (purum p.a., ≥ 30%) to 990 µl bi-distilled water to create a 100 mM stock solution to be used within 12 hr.
- Dilute the stock solution from step 1.2.3 to 50 µM in EBSS right before the experiment.
- Incubate the cell spheroids via the microfluidic system during measurement with hydrogen peroxide 50 µM in EBSS for oxidation of the redox sensor.
- Experiment 3: Initiation and labeling of cellular necrosis within spheroids
- Seed about 400 cells (e.g., U251-MG-L106-Grx1-roGFP2 glioblastoma cells) in agarose gel–coated wells (procedure described in steps 1.1.1–1.1.3) of a 96-well plate in DMEM culture medium supplemented with 10% FCS, 1% penicillin/streptomycin and hygromycin B (150 mg/ml).
- Grow spheroids for 5–7 days up to a diameter of about 200–300 µm in an incubator at 5% CO2 and 37 °C.
- Incubate the cell spheroids with the mitochondrial electron transport inhibitor rotenone for 3 hr at a concentration of 1 µM in culture medium to induce cellular necrosis. Additionally, co-incubate with a green cytotoxicity dye for 3 hr at a concentration of 1 µl in 1 ml culture medium to label necrotic cells.
- Wash the cell spheroids with culture medium or EBSS prior to microscopy.
2. Light Sheet Adjustment
NOTE: For achieving best results it has to be ensured that the beam waist of the light sheet is in the focus plane of the objective lens. The position of the beam waist can only be aligned by observing the light sheet in vertical position with respect to the capillary (see steps 2.5 - 2.8).
- Use an inverted microscope and mount the SPIM excitation module, as described in Ref. 16, to the baseplate of the positioning table (see Figure 1A).
- Equip the microscope with a 10X/0.3 or 20X/0.5 microscope objective lens and an appropriate long-pass filter (e.g., λ ≥ 515 nm) in the detection path of the microscope.
- Mount an integrating camera to the detection port of the microscope.
- Use a parallel collimated beam of a laser or a laser diode with an excitation wavelength of preferentially 470 nm and apply it to the SPIM module.
- Rotate the cylindrical lens by 90 degrees, which transfers the light sheet into a vertical position for an axial adjustment of the beam waist of the light sheet.
- Place a capillary containing a liquid with a fluorescent dye in the sample holder.
- Attach the sample holder with the capillary to the positioning table of the microscope and align the position of the capillary such that it is centered and the beam waist of the light sheet is in focus.
- Adjust the beam waist by variation of the axial position of the cylindrical lens itself. Turn back the lens after adjustment.
3. Cell Spheroid Application and Microscope Feed Synchronization
- Place the cell spheroids separately or in groups within rectangular borosilicate glass capillaries16 with an inner cross section of 600 µm x 600 µm and a wall thickness of 120 µm. Two techniques for the application of a cell spheroid have proven to be successful.
- Take the empty capillary upright with your thumb and middle finger and seal the upper opening with your forefinger. Bring the lower opening close to the cell spheroid in its surrounding liquid (EBSS or culture medium). Release the forefinger from the upper opening. Liquid with the cell spheroid in it will be soaked in immediately by capillary forces. Adjust the position of the spheroid within the filled capillary by gravitation in respective upright position of the capillary.
- Alternatively, apply the cell spheroid to the capillary via pipetting. Take care that the opening of the tip of the pipette is, on the one hand, big enough for the spheroid size and, on the other hand, less or equal in size to the inner diameter of the capillary.
- Place the capillary with the spheroid in it in a special sample holder for microscopy.
- Attach the sample holder with the capillary to the positioning table of the microscope and align the position of the cell spheroid such that it is centered and focused.
- Set the matching between the light sheet and the focal plane of the microscope objective lens in the detection path by adjusting the position of the reflection mirror and the cylindrical lens (see adjustment screw in Figure 1B).
- Adjust the micrometer screw (see Figure 1B) according to the refraction indices and the numerical aperture of the objective lens to compensate the fishtank effect.
NOTE: Different refraction indices of the sample and the media surrounding the objective lens lead to a difference between the shift of the objective lens and the shift of the focal plane. Since the light sheet is moved by the objective turret, the mismatch between the shift of the light sheet (corresponding to the shift of the objective lens) and the shift of the focal plane is compensated by a lever arm (see Figure 1B). The micrometer screw is used to adapt the distance between the upper and the lower pin to adjust the shift of the light sheet according to the numerical aperture of the objective lens and the refraction indices.
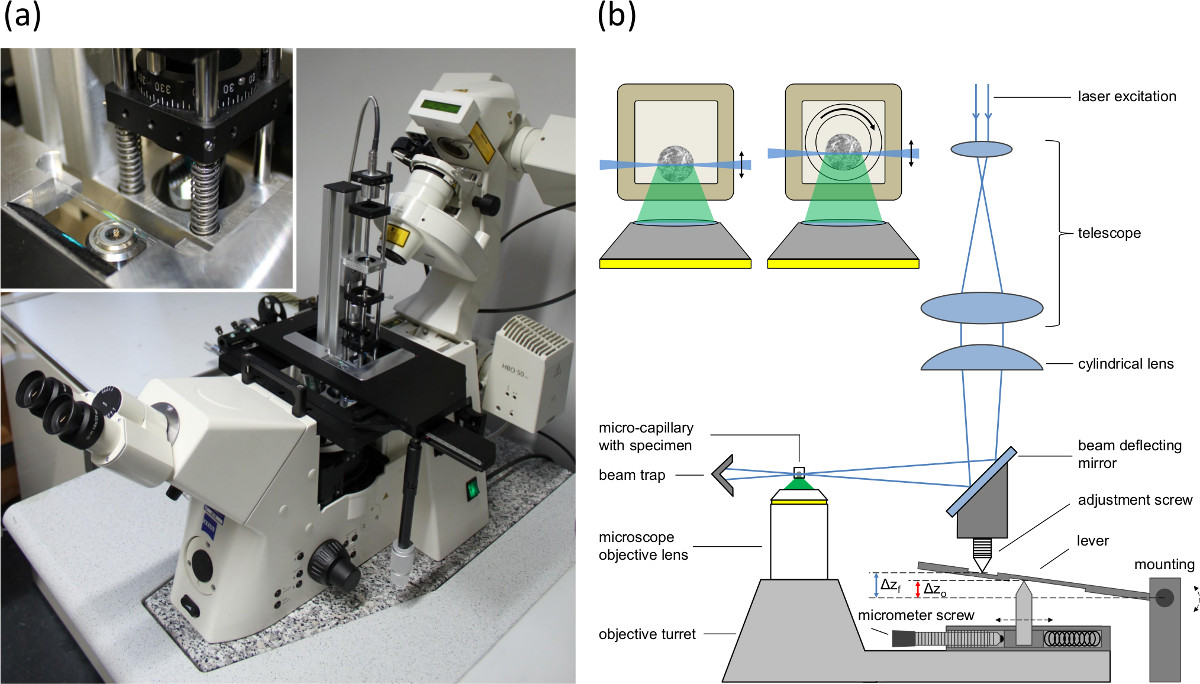
Figure 1. (A) Picture of the single plane illumination module mounted to the baseplate of the positioning table of an inverted microscope, (B) setup of the single plane illumination module and the microscope feed synchronization to compensate the lifting-mismatch (fishtank effect) between focal plane and illumination plane. Δzo indicates the shift of the objective lens and Δzf the shift of the focal plane and the light sheet. The inlay shows cross sections of spheroid containing capillaries. Left: rectangular capillary with inner diameter of 600 µm (wall 120 µm). Right: Setup used for rotation. An inner round capillary (inner diameter 400 µm, outer diameter 550 µm) is rotated within the outer rectangular capillary. The space between the two capillaries is filled with an immersion fluid.
4. Measurement in Dynamic Liquid Environment
NOTE: The previous protocol is used for static incubation prior to a measurement where the cell spheroid has previously been incubated and then held in place in the capillary by simple gravitation. There is no need for further fixation. However, for measurements in flowing media where a dynamic incubation and, therefore, a dynamic environment is desirable to measure uptake kinetics, one can accomplish this sub-protocol. Steps 4.5–4.9 are critical to avoid air bubbles reaching the sample.
- Fill the capillary with FCS for 30 min to support later cellular adhesion of a cell spheroid to the inner glass surface.
- Let the remaining FCS coating in the depleted capillary dry out for at least 12 hr.
- Introduce the cell spheroid to the FCS coated capillary as described in step 3.2.
- Leave the capillary in the incubator at 5% CO2 and 37 °C for additional 2–4 hr to cause cellular adhesion of the spheroid.
- Set up the afflux part of the microfluidic system (see Figure 2). Use a peristaltic pump to fill the afflux of the tubing (inner diameter: 0.89 mm) with culture medium containing the fluorescent dye, drug or agent.
- Connect the afflux part of the microfluidic system to a bubble trap (see Figure 2).
- First clamp the capillary bubble-free to the afflux tubing and then clamp the other side of the capillary to the drain tubing.
- Tune the liquid temperature of the water bath to the desired value (e.g., 37 °C).
- Adjust the peristaltic pump to the desired pump velocity (e.g., flow rate: 9 µl/min; pump velocity in the capillary: 25 mm/min).
- Collect the pumped liquid either in a recipient (open-loop setup), feed it back to its source (closed-loop setup) or feed it directly into the tubing of the peristaltic pump (tight closed-loop setup).
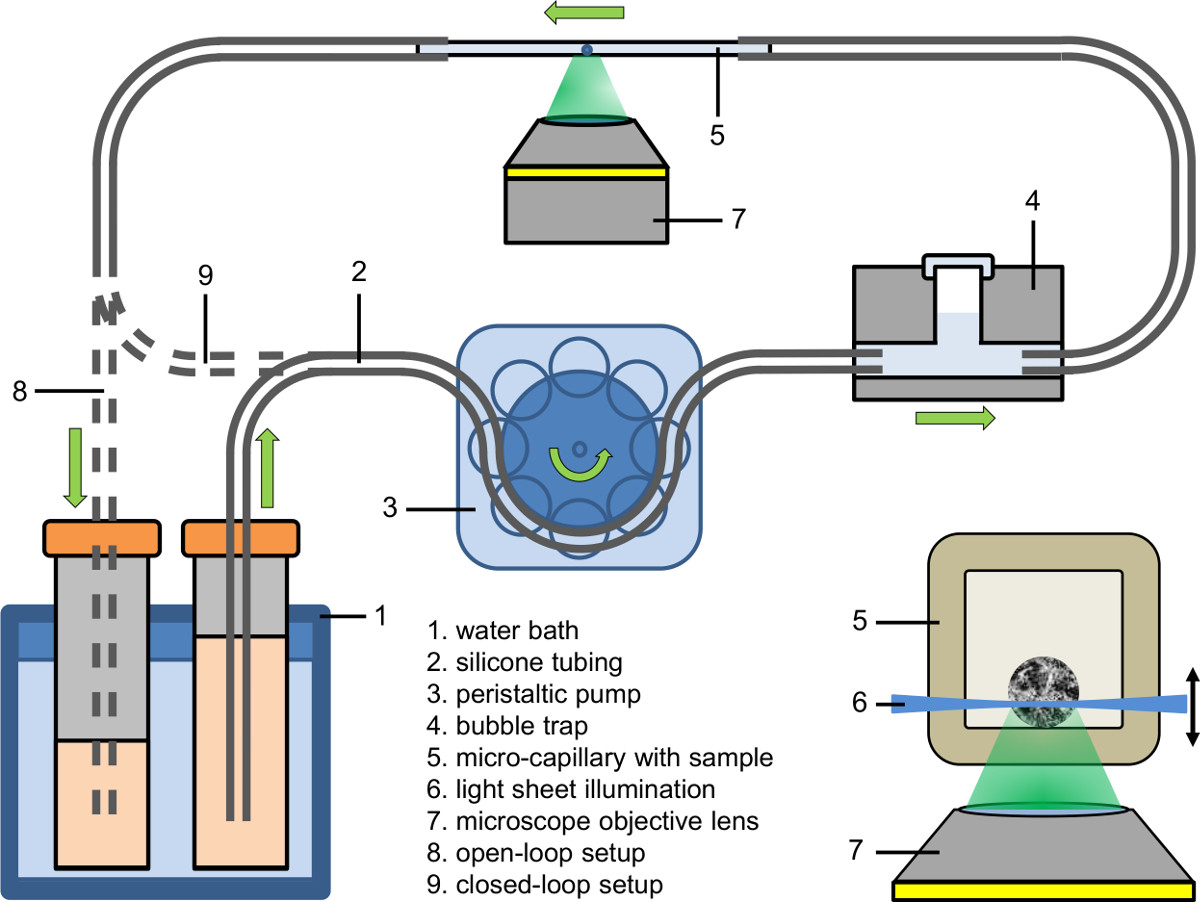
Figure 2. Microfluidic setup (open-loop and tight closed-loop); inlay: Illumination of a spheroid within a micro-capillary using light sheet based fluorescence microscopy coupled to an inverted microscope.
5. Data Acquisition and Analysis
- Set the laser power and the integration time for the image acquisition.
- Take care that the parameters defined in step 5.1 do not exceed light dose values about 50–100 J/cm2 for native cells or 10–20 J/cm2 for cells incubated with a fluorescent dye or transfected with a fluorescent protein encoding plasmid to avoid phototoxicity (for details see Ref. 12).
- Set the increment for the z-stack to ∆z = 5–10 µm which is preferable for 3-dimensional data analysis as the light sheet thickness is about 10 µm.
- Perform measurements of single images or z-stacks by variation of the focal plane within the cell spheroid.
Results
Experiment 1: Cell spheroids incubated with a chemotherapeutic drug
A z-stack scan of a previously incubated MCF-7 cell spheroid (8 µM doxorubicin, 6 hr) is depicted in Figure 3. It gives detailed information about the cellular uptake and distribution of doxorubicin17 and its degradation product18,19. Within the outer cell layer of the spheroid red fluorescent doxorubicin is mainly localized in the nucleus, whereas in inner...
Discussion
The present manuscript describes a light sheet or single plane illumination microscopy (SPIM) device which is optimized for 3-dimensional cell systems, e.g., multi-cellular tumor spheroids (MCTS). Three exemplary applications include (1) uptake of a cytostatic drug and its partial conversion to a degradation product (whose contribution to chemotherapeutic efficacy still remains to be evaluated), (2) measurements of the redox state by use of a genetically encoded glutathione sensor upon addition of an oxidizing a...
Disclosures
No conflicts of interest declared.
Acknowledgements
This project was funded by the Land Baden-Württemberg as well as by the European Union, Europäischer Fonds für die Regionale Entwicklung. The authors thank Rainer Wittig (ILM Ulm) for providing the U251-MG-L106-Grx1-roGFP2 cell line and Claudia Hintze for skillful technical assistance.
Materials
| Name | Company | Catalog Number | Comments |
| Microtiter plate | Orange Scientific | 4430100 | For cell spheroid growing |
| Agarose | Carl Roth GmbH | 3810.1 | For cell spheroid growing |
| MCF-7 cell line | CLS Cell Lines Service GmbH | 300273 | Cell line |
| U251-MG-L106 cell line | Cell line | ||
| DMEM | Biochrom AG (Merck Millipore) | FG0435 | Culture medium |
| DMEM/Ham's F-12 | Biochrom AG (Merck Millipore) | FG4815 | Culture medium |
| FCS | Biochrom AG (Merck Millipore) | S0615 | Cell culture supplement |
| Penicillin/streptomycin | Biochrom AG (Merck Millipore) | A 2213 | Antibiotics |
| Hygromycin B | PAA Laboratories | P02-015 | Antibiotic |
| EBSS | Sigma-Aldrich Inc. | E3024 | Cell culture supplement |
| Doxorubicin | Sigma-Aldrich Inc. | D1515 | Fluorescent dye (CAUTION: acute toxicity) |
| Green cytotoxicity dye | Promega GmbH | G8742 | CellTox - fluorescent cytotoxicity dye |
| Rotenone | Sigma-Aldrich Inc. | R8875 | Cellular inhibitor (CAUTION: acute toxicity) |
| Hydrogen peroxide (H2O2) | Sigma-Aldrich Inc. | 95302 | Reagent for oxidation (CAUTION: acute toxicity) |
| Capillary | VitroCom | 8260-050 | Sample preparation |
| Microscope | Carl Zeiss Jena | Axiovert 200M | |
| AxioCam MRc CCD-camera | Carl Zeiss MicroImaging GmbH | 426508-9901-000 | CCD-camera |
| AxioVision data aquisition software | Carl Zeiss MicroImaging GmbH | version 4.8.2. | |
| Laser diode | Pico Quant GmbH | LDH-P-C-470 | Used with driver PDL800-B |
| Peristaltic pump | Ismatec Labortechnik | MS-1 Reglo | Re-calibrated to reduce the minimum pump speed by 1/10 |
| Silicone tubes | IDEX Health & Science GmbH | TYGON R3607 |
References
- Pawley, J. . Handbook of biological confocal microscopy. , (1990).
- Webb, R. H. Confocal optical microscopy. Rep. Prog. Phys. 59, 427-471 (1996).
- Denk, W., Strickler, J. H., Webb, W. W. Two-photon laser scanning fluorescence microscopy. Science. 248 (4951), 73-76 (1990).
- König, K. Multiphoton microscopy in life sciences. J. Microsc. 200 (2), 83-104 (2000).
- Neil, M. A., Juskaitis, R., Wilson, T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt. Lett. 22 (24), 1905-1907 (1997).
- Gustafsson, M. G. L., et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94 (12), 4957-4970 (2008).
- Huisken, J., Swoger, J., del Bene, F., Wittbrodt, J., Stelzer, E. H. K. Optical sectioning deep inside live embryos by SPIM. Science. 305 (5686), 1007-1009 (2004).
- Huisken, J., Stainier, D. Y. R. Selective plane illumination microscopy techniques in development biology. Development. 136 (12), 1963-1975 (2009).
- Santi, P. A. Light sheet fluorescence microscopy: a review. J. Histochem. Cytochem. 59 (2), 129-138 (2011).
- Kunz-Schughart, L. A., Freyer, J. P., Hofstaedter, F., Ebner, R. The use of 3-D cultures for high throughput screening: the multi-cellular spheroid model. J. Biomol. Screen. 9 (4), 273-285 (2004).
- Schneckenburger, H., et al. Multi-dimensional fluorescence microscopy of living cells. J. Biophotonics. 4 (3), 143-149 (2011).
- Schneckenburger, H., et al. Light exposure and cell viability in fluorescence microscopy. J. Microsc. 245 (3), 311-318 (2012).
- Ritter, J. G., Veith, R., Veenendaal, A., Siebrasse, J. P., Kubitschek, U. Light sheet microscopy for single molecule tracking in living tissue. PLoS One. 5 (7), e11639:1-e11639:7 (2010).
- Mertz, J., Kim, J. Scanning light-sheet microscopy in the whole mouse brain with HiLo background rejection. J. Biomed. Opt. 15 (1), 016027 (2010).
- Fahrbach, F. O., Rohrbach, A. A line-scanned light-sheet microscope with phase shaped self-reconstructing beams. Opt. Express. 18 (23), 24229-24244 (2010).
- Bruns, T., Schickinger, S., Wittig, R., Schneckenburger, H. Preparation strategy and illumination of 3D cell cultures in light-sheet based fluorescence microscopy. J. Biomed. Opt. 17 (10), 101518 (2012).
- Ma, H. L., et al. Multicellular tumor spheroids as an in vivo-like tumor model for three-dimensional imaging of chemotherapeutic and nano material cellular penetration. Mol. Imaging. 11 (6), 487-498 (2012).
- Weber, P., Wagner, M., Schneckenburger, H. Cholesterol dependent uptake and interaction of doxorubicin in MCF-7 breast cancer cells. Int. J. Mol. Sci. 14 (4), 8358-8366 (2013).
- Hovorka, O., et al. Spectral analysis of doxorubicin accumulation and the indirect quantification of its DNA intercalation. Eur. J. Pharm. Biopharm. 76 (3), 514-524 (2010).
- Schickinger, S., Bruns, T., Wittig, R., Weber, P., Wagner, M., Schneckenburger, H. Nanosecond ratio imaging of redox states in tumor cell spheroids using light sheet-based fluorescence microscopy. J. Biomed. Opt. 18 (12), 126007 (2013).
- Lippert, H., Wald, M., Radt, B. Optical arrangement for the production of a light sheet. U.S. Patent. , (2010).
- Lorenzo, C., et al. Live cell division dynamics monitoring in 3D large spheroid tumor models using light sheet microscopy. Cell Division. 6 (22), 1-8 (2011).
- Bruns, T., Schneckenburger, H., Schickinger, S. Sample holder for rotation of three-dimensional specimens in microscopy. European Patent Application. , (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved