Method Article
Identification of Kinesin-1 Cargos Using Fluorescence Microscopy
In This Article
Summary
Here, a protocol is presented to identify Kinesin-1 cargos. A motorless mutant of the Kinesin-1 heavy chain (KIF5B) aggregates in the cytoplasm and induces aggregation of its cargos. Both aggregates are detected under fluorescence microscopy. A similar strategy can be employed to identify cargos of other motor proteins.
Abstract
Fluorescence microscopy is employed to identify Kinesin-1 cargos. Recently, the heavy chain of Kinesin-1 (KIF5B) was shown to transport the nuclear transcription factor c-MYC for proteosomal degradation in the cytoplasm. The method described here involves the study of a motorless KIF5B mutant for fluorescence microscopy. The wild-type and motorless KIF5B proteins are tagged with the fluorescent protein tdTomato. The wild-type tdTomato-KIF5B appears homogenously in the cytoplasm, while the motorless tdTomato-KIF5B mutant forms aggregates in the cytoplasm. Aggregation of the motorless KIF5B mutant induces aggregation of its cargo c-MYC in the cytoplasm. Hence, this method provides a visual means to identify the cargos of Kinesin-1. A similar strategy can be utilized to identify cargos of other motor proteins.
Introduction
Kinesin-1 is a motor protein that mediates anterograde transport of its cargos1,2. It is a heterotetramer of the two subunits of Kinesin Light Chain 1 (KLC1) and two subunits of Kinesin Heavy Chains (KHCs). KIF5B1, a KHC, contains a motor domain at its N-terminal, which hydrolyzes ATP and converts the chemical energy to mechanical energy for movement along microtubules. Its C-terminal region contains the dimerization domain that interacts with KLC1, which associates with cargos. Kinesin-1 transports cargos such as vesicles, organelles and mRNAs3,4. Recently, KIF5B was shown to transport the nuclear transcription factor c-MYC for proteosomal degradation in the cytoplasm5. Three methodologies (chemical inhibitor, siRNA/ shRNA and dominant negative mutant) were used to inhibit Kinesin-1 function. They all induced aggregation of c-MYC in the cytoplasm. For the last methodology, c-MYC was only affected by the dominant negative mutant of KIF5B, but not by that of another related KIF5A motor protein, suggesting that the mutant does not exert general effects on the intracellular components (like microtubule disruption) or on protein aggregation. The dominant negative mutant of KIF5B also did not affect another transcription factor, suggesting that it does not exert general effects on transcription factors. Rather, it suggests that the dominant negative mutant exerts specific effects on its cargos.
The use of dominant negative mutants is common in the field of motor proteins. Similar motorless mutants of kinesins and myosins were used previously. They were mainly used to demonstrate the effects of the mutants on the subcellular localizations of their cargos or on cellular functions6-12. Less emphasis was put on the spatial relationship between the mutants and the cargos affected by them. However, in some incidences, the mutants were observed to co-localize with their cargos6,10.
The interaction between KIF5B and its associated proteins was previously confirmed by the in vivo yeast two-hybrid assay and biochemical pull-down assays such as co-immunoprecipitation and in vitro pull-down assays13-16. In this article, an additional visual method using fluorescence microscopy is described to identify KIF5B cargo proteins. The method makes use of a motorless KIF5B mutant that acts as a dominant negative mutant. It aggregates in the cytoplasm and induces aggregation of its cargos.
The tagging of wild-type and motorless KIF5B mutant with the fluorescent protein tdTomato17 enables their visualization by fluorescence microscopy. The tagged KIF5B proteins can be co-expressed with a candidate protein fused to a different fluorescent protein with spectral properties suitably separated from the KIF5B tag. The tagged proteins are observed directly in live cells under fluorescence microscopy. Induction of aggregation of the candidate protein by the motorless KIF5B mutant will confirm that the candidate protein is an in vivo cargo of KIF5B. Furthermore, the tdTomato-tagged KIF5B proteins can be expressed alone in the cells to study their effects on the endogenous cargo proteins. Later, immunofluorescence microscopy is conducted in which the transfected cells are fixed and stained with a specific antibody against the endogenous candidate protein, followed by an appropriate secondary antibody conjugated with a fluorescent dye. In this case, the endogenous candidate protein at its physiological level is studied. Similar motorless mutants of other motor proteins can be prepared to identify their cargos.
Protocol
1. Cloning of the tdTomato-tagged Wild-type and Motorless KIF5B Proteins
- Amplify the cDNAs for the human wild-type and motorless KIF5B proteins using the primers in Table 1, Taq DNA polymerase (5 units for 100 μl), dNTP mix (2 mM for each deoxynucleotide) and its 10X buffer for 30 cycles. Each cycle consists of a denaturation step (95 °C for 30 sec), an annealing step (45 °C for 30 sec) and an extension step (72 °C for 3 min).
- Extract the amplified DNA product with equal volume of phenol/ chloroform (1:1).
Note: Phenol is combustible and can cause burns. Chloroform is hazardous. Avoid direct contact with them and use them under a chemical fume hood. Alternatively, PCR products can be purified by various kits.- Spin at 18,000 x g in a microcentrifuge at room temperature for 1 min. Transfer the aqueous solution to a new tube and extract with an equal volume of chloroform.
- Spin at 18,000 x g in a microcentrifuge at room temperature for 0.5 min. Transfer the aqueous solution to a new tube.
- Mix the aqueous solution with one tenth volume of 3 M Na-acetate and two volumes of absolute ethanol.
- Spin at 18,000 x g in a microcentrifuge at room temperature for 5 min. Discard the aqueous solution.
- Wash the DNA pellet with two volume of 75% ethanol. Discard the aqueous solution and air dry the DNA pellet at room temperature for 5 min. Resuspend the DNA pellet in 34 μl water.
- Digest the amplified products in a final volume of 40 μl with restriction enzymes SalI (10 units) and BamHI (10 units) and 4 μl of their 10X buffer at 37 °C for two hr. Resolve the digested products in an agarose gel (1.0% at 100 V) containing ethidium bromide (0.2 μg/ml).
Note: Ethidium bromide is a potent mutagen and can be absorbed through skin. Therefore, it is important to avoid direct or indirect contact with ethidium bromide. - Cut out the correct bands for purification by columns. Weigh the agarose gel containing the DNA fragment. Then dissolve it in the solubilization buffer (300 μl for 0.1 g) at 37 °C for about 20 min.
- Add the resulted solution to a column and spin in a microcentrifuge at room temperature for 5 sec. Discard the flow-through.
- Wash the column with 0.5 ml solubilization buffer by repeating the step 1.5. Wash the column again with 0.75 ml wash buffer by repeating the step 1.5. Spin the column for 2 min to get rid of the remaining wash buffer.
- Elute the DNA fragment with 50 μl water by repeating step 1.5. Estimate the concentration of the DNA fragment by running an aliquot of it against a DNA size ladder in an agarose gel (1.0% at 100 V) containing ethidium bromide (0.2 μg/ml).
- Ligate the purified products (about 100 ng) with the ptdTomato-C1 vector17 (about 100 ng) previously digested with the same restriction enzymes in 10 μl using T4 DNA ligase (2,000 units) and 1 μl 10X ligation buffer at room temperature for 16 hr.
Note: The wild-type and motorless mutant contains amino acids from 2 to 963 and 584 to 963, respectively. Previously, a larger motorless KIF5B mutant was used to identify its function9. A smaller motorless KIF5B mutant was used for the present studies to increase its expression levels.
2. Immunofluorescence Microscopy
- For live-cell or indirect fluorescence imaging with magnification at or below 40X, seed 0.2-0.3 million HeLa cells in 1 ml complete medium [Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS)] in each well of a six-well plate. Grow at 37 °C with 5 % CO2.
- For indirect immunofluorescence studies with magnifications above 40X, wash cover glasses (18 mm x 18 mm; 1.5 thickness) in absolute ethanol briefly and air-dry them in the wells of a six-well plate, inside a biosafety cabinet to avoid contamination.
- Seed 0.2-0.3 million cells in 1 ml complete DMEM medium in each well of a six-well plate.
- Preparation of Transfection Complex
- The next day, inside a biosafety cabinet, dilute 0.6 µg (total) of the expression plasmid for tdTomato-tagged wild-type or motorless KIF5B in the presence or absence of an expression plasmid for a cargo candidate protein (pTagCFP-c-MYC5) with 0.1 ml transfection medium in a 1.6 ml microcentrifuge tube.
Note: The vector pTagCFP-c-MYC expresses the TagCFP-tagged c-MYC. - Add 1.8 µl of the transfection reagent to 0.1 ml transfection medium in another 1.6 ml microcentrifuge tube. Typically, prepare a master mix of diluted transfection reagent enough for 12 samples.
- Incubate for 5 min, at room temperature.
- Add the diluted 0.1 ml transfection reagent to the diluted 0.1 ml DNAs. Mix the contents by inverting the tubes. Spin the tubes for 5 sec in a microcentrifuge.
- Incubate for 45 min, at room temperature.
- The next day, inside a biosafety cabinet, dilute 0.6 µg (total) of the expression plasmid for tdTomato-tagged wild-type or motorless KIF5B in the presence or absence of an expression plasmid for a cargo candidate protein (pTagCFP-c-MYC5) with 0.1 ml transfection medium in a 1.6 ml microcentrifuge tube.
- Washing of Cells
- Wash the seeded cells three times with phosphate buffered saline (PBS) during the incubation period for the formation of the transfection complex.
- Replace the medium of the seeded cells in each well with 0.8 ml pre-warmed transfection medium. Return the plates to the incubator at 37 °C.
- Transfection of Cells
- After incubation of 45 min, add 0.2 ml of the DNA/transfection reagent complex (prepared in step 2.3) dropwise to each well of the plate.
- Rock the 6 well plate gently for 5 sec, before they are returned to the incubator at 37 °C.
- After 6-8 hr, replace the medium with the complete DMEM medium.
- For Live-cell Fluorescence Imaging18
- After an additional 16 hr of incubation at 37 °C, add the DNA intercalating stain Hoechst 33342 to the medium to a final concentration of 1 µM.
Note: Hoechst 33342 is a cell-permeable, DNA intercalating dye that stains the cell nuclei. - Incubate the transfected cells for 10 min in the incubator at 37 °C before examining them for the expression of fluorescent proteins by fluorescent microscopy using a 40X objective. The filter sets for DAPI, CFP, FITC and Cy3 are (Ex350 nm/ Em460 nm), (Ex436 nm/ Em480 nm), (Ex470 nm/ Em525 nm) and (Ex545 nm/ Em605 nm), respectively.
Note: If the background is high due to auto-fluorescence of the complete DMEM medium, medium without phenol red is used.
- After an additional 16 hr of incubation at 37 °C, add the DNA intercalating stain Hoechst 33342 to the medium to a final concentration of 1 µM.
3. For Indirect Immunofluorescence Studies
- Preparation of Paraformaldehyde Solution
- Weigh paraformaldehyde powder (4 g per 100 ml PBS) and add it to PBS.
Note: Paraformaldehyde is a flammable solid and a potential cancer hazard. It irritates eyes, respiratory system and skin. Therefore, it is important to avoid contact with or inhalation of paraformaldehyde. - Add NaOH to the solution (150 µl 10 N NaOH/100 ml).
- Keep the solution at 37 to 42 °C for 2-3 hr with occasional shaking.
- Adjust pH to 7.0 by adding glacial acetic acid to the solution (about 75 µl/ 100 ml) after the paraformaldehyde powder is dissolved.
- Weigh paraformaldehyde powder (4 g per 100 ml PBS) and add it to PBS.
- Target Proteins Staining
- After further incubation for 16 hr at 37 °C, wash the transfected cells at room temperature once with PBS by swirling the six-well plate for 5 sec.
- Fix the cells with 1 ml of freshly prepared, 4% paraformaldehyde solution per well. Incubate at room temperature for 30 min.
- Wash the cells with PBS once by swirling for 5 sec the six-well plate. Discard the solution.
- Incubate the cells with 1 ml of 0.1% Triton-X 100 (in PBS) at room temperature for 30 min. The detergent Triton-X 100 will permeabilize the cell membrane to allow access of antibodies to their intracellular targets.
- Wash the cells with PBS four times by swirling the six-well plate for 5 sec each time. Discard the solution after each wash.
- Incubate the cells with 1 ml of primary antibodies (c-MYC rabbit antibody or p53 mouse antibody; 0.1 µg/ml) in a 10% FBS in PBS solution at room temperature by rocking for 4 hr.
- Wash with PBS four times by swirling the six-well plate for 5 sec each time. Discard the solution after each wash.
- Incubate with 1 ml of fluorescent dye-conjugated secondary antibodies (Alexa Fluor 488-conjugated anti-rabbit or anti-mouse IgG antibody; 0.5 µg/ml) in 10% FBS in PBS at room temperature in the dark by rocking for 2 hr.
- Wash with PBS four times by swirling the six-well plate for 5 sec each time. Discard the solution after each wash.
- Nuclear Staining and Mounting
- Incubate the cells with the 1 ml of DNA intercalating dye 4',6-diamidino-2-phenylindole (DAPI; 0.5 µg/ml) in PBS at room temperature, in dark for 10 min. Proceed to step 3.4 when no cover glasses are utilized.
Note: DAPI is a DNA intercalating dye that stains the cell nuclei. - Apply 10 μl of mounting solution onto each microscope slide. Mount each cover glass over the mounting solution on a microscope slide.
- Incubate at room temperature in the dark overnight.
- Seal the edges of the cover glasses with nail polish. Place in the dark inside a fume hood overnight to remove the nail polish fumes.
- Incubate the cells with the 1 ml of DNA intercalating dye 4',6-diamidino-2-phenylindole (DAPI; 0.5 µg/ml) in PBS at room temperature, in dark for 10 min. Proceed to step 3.4 when no cover glasses are utilized.
- Fluorescence Microscopy19
- Next day, examine the cells by fluorescent microscopy using a 40X objective. The filter sets for DAPI, CFP, FITC and Cy3 are (Ex350 nm/ Em460 nm), (Ex436 nm/ Em480 nm), (Ex470 nm/ Em525 nm) and (Ex545 nm/ Em605 nm), respectively.
Results
Exogenously expressed wild-type KIF5B appeared homogeneously in the cytoplasm, while c-MYC appeared mainly in the nucleus (Figure 1A). However, the motorless KIF5B mutant formed aggregates in the cytoplasm (Figure 1A). Aggregation of the motorless KIF5B induced the aggregation of c-MYC. The percentage of cells expressing aggregated TagCFP-tagged c-MYC in the cells expressing wild-type KIF5B was low. However, it was significantly higher when the motorless KIF5B was co-expressed (Figure 1A). The observed co-localization of mutant KIF5B with c-MYC (Figure 1) suggests that KIF5B regulates the subcellular localization of c-MYC and c-MYC is a cargo of Kinesin-1. Negative controls in which the constructs were expressed alone are included in the lower panel of Figure 1A. The results show that there was no significant bleed-through of fluorescence emission. The motorless KIF5B also induced aggregation of the endogenous c-MYC (Figure 1B) and the transcription factor p53 (Figure 2), indicating that c-MYC and p53 are both the cargos of Kinesin-1 and KIF5B regulates the subcellular localization of both endogenous proteins. Together with the results that the Kinesin-1 inhibitor rose bengal lactone (RBL)20 induces formation of high molecular weight species for both c-MYC and p535, p53 is likely a cargo of Kinesin-1. It is interesting to note that the nuclear transcription factor p53, like c-MYC, is also exported from the nucleus to the cytoplasm for proteasomal degradation21,22. The movement of transcription factors by KIF5B appears to be specific because the expression of the motorless KIF5B mutant did not affect the subcellular localization c-Fos or cause it to aggregate (Figure 3). The above data demonstrate that the method employed in this publication allows for the high-specificity identification of Kinesin-1 cargos. A similar strategy may be applied to other motor proteins as well.
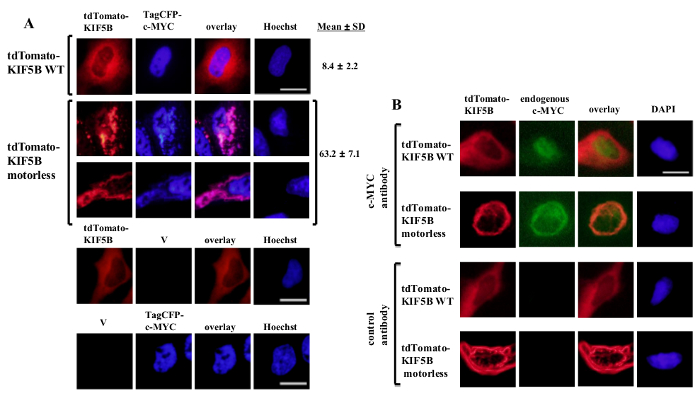
Figure 1: Expression of the motorless KIF5B mutant induces c-MYC aggregation in the cytoplasm. (A) HeLa cells were transfected with tdTomato-tagged wild-type (WT) KIF5B (red) and TagCFP-tagged c-MYC (blue). tdTomato-tagged WT KIF5B appeared mainly in the cytoplasm, while TagCFP-tagged c-MYC appeared mainly in the nucleus. However, the tdTomato-tagged motorless KIF5B mutant (red) formed filamentous or punctate aggregates in the cytoplasm. Expression of the motorless KIF5B induced the aggregation of c-MYC. The mutant KIF5B and c-MYC co-localized together (pink). Percentages of cells (%) indicating aggregates of CFP-c-MYC (%) are shown. The results are shown as mean±SD (N = 3). Negative controls with expression of tdTomato-tagged KIF5B proteins or TagCFP-tagged c-MYC along with empty vectors (V) are shown in the lower panel. (B) Similar results were obtained with the endogenous c-MYC (green) when tdTomato-tagged WT or motorless KIF5B proteins (red) were expressed. The tdTomato-tagged motorless KIF5B mutant formed aggregates in the cytoplasm and induced aggregation of the endogenous c-MYC. Both kinds of aggregates co-localized together in the cytoplasm (orange). The nuclei were stained with the dye Hoechst 33342 or DAPI. Scale bar; 20 μm. Please click here to view a larger version of this figure.

Figure 2: Expression of the motorless KIF5B mutant induces endogenous p53 aggregation in the cytoplasm. In HeLa cells, exogenous tdTomato-tagged wild-type (WT) KIF5B (red) appeared mainly in the cytoplasm, while the endogenous p53 (green) appeared mainly in the nucleus. In contrast, the tdTomato-tagged motorless KIF5B mutant (red) formed aggregates in the cytoplasm. Expression of the motorless KIF5B induced the aggregation of p53, resulting in the co-localization of KIF5B and p53 in the cytoplasm (yellow). The experiment was performed three times. The nuclei were stained with the dye DAPI. Scale bar; 20 μm. Please click here to view a larger version of this figure.
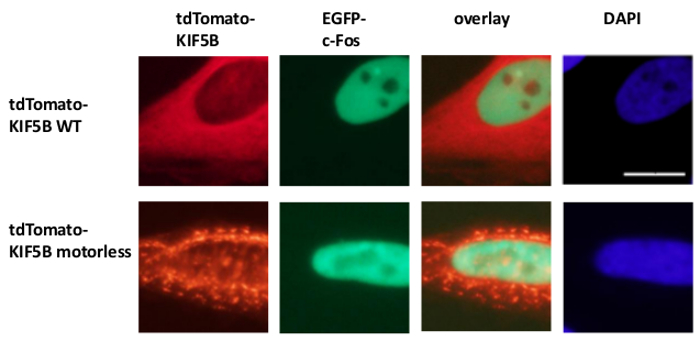
Figure 3: Expression of the motorless KIF5B mutant does not induce aggregation of c-Fos in the cytoplasm. tdTomato-tagged wild-type (WT) KIF5B (red) appeared mainly in the cytoplasm of HeLa cells, while EGFP-c-Fos (green) appeared mainly in the nucleus. On the contrary, the tdTomato-tagged motorless KIF5B mutant (red) formed aggregates in the cytoplasm. Expression of the motorless KIF5B did not induce the aggregation of c-Fos. The experiment was performed four times. The nuclei of the cells were stained with the dye DAPI. Scale bar; 20 μm. Please click here to view a larger version of this figure.
| common reverse primer | 5’-AGAGGATCCTTACACTTGTTTGCCTCCTC-3’ |
| wild-type KIF5B forward primer | 5’- AGAGTCGACGCGGACCTGGCCGAGTGCAACATCAAAGT-3’ |
| motorless KIF5B forward primer | 5’- AGAGTCGACGATGAAGAGTTCACTGTTGC-3’ |
Table 1: Primer sequences for cloning the wild-type and motorless KIF5B proteins.
Discussion
The method described utilizes the properties of the motorless KIF5B mutant, which lacks the ability to move along the microtubules, but retains the ability to form dimers with the wild-type KIF5B and, thereby, allow the tetrameric protein to interact with the same cargo proteins as the wild-type KIF5B. Motorless KIF5B, therefore, acts as a dominant negative mutant and forms mislocalized aggregates with its cargos. This method is proven to identify the Kinesin-1 cargo c-MYC (Figure 1)5. In this article, the same motorless KIF5B mutant was used to identify p53 as another potential cargo of Kinesin-1 (Figure 2). This shows that the methodology is feasible to identify other cargos of Kinesin-1. Furthermore, the specificity of the mutant is provided by the lack of effect of the mutant on the negative control protein c-Fos (Figure 3).
In this protocol, the tdTomato-tagged wild-type or motorless KIF5B protein is coexpressed with another fluorescent protein-tagged candidate cargo protein. In this case, live-cell fluorescence microscopy and imaging are performed. The formation of the aggregates can be traced by time-lapse imaging. Alternatively, the tdTomato-tagged protein is expressed alone and the candidate cargo protein at its physiological levels is visualized by indirect immunofluorescence microscopy using specific antibodies. The fluorescent protein tdTomato is chosen for its brightness and photostability17. If the background is high due to auto-fluorescence of the complete DMEM medium, medium without phenol red is used.
The motorless KIF5B mutant forms dimers with the wild-type KIF5B stoichiometrically. Therefore, it is critical to express sufficient amount of motorless KIF5B mutant to form dimers with the wild-type counterpart to inhibit its function and to form aggregates. To address this issue, optimization of expression of the motorless mutant is essential. It is achieved by using a small motorless mutant containing the dimerization domain. In addition, optimization of the appropriate transfection reagent and protocol is also necessary. The duration of incubation of HeLa cells with the DNA/ transfection reagent was optimized in HeLa cells for expression of exogenous proteins and cell viability. The incubation duration may have to be optimized for other cell lines.
For magnifications between 4X to 40X, no cover glasses are required. Cells can be directly examined in the wells. Therefore, in this case, the protocol is inexpensive and convenient. For magnifications above 40X, cells are grown on cover glasses in the wells and, after staining, are mounted on microscope slides for examination under oil-immersion objectives.
Observation of aggregates of the motorless KIF5B protein is limited by the size of the cytoplasm. It is easy to detect the aggregates in many mammalian cells. However, it is relatively difficult to observe the aggregates in neuronal cells when the volume of the cytoplasm is small around the nuclei and in neurites.
The protocol is used to show the association between KIF5B and its cargos in addition to the in vivo yeast two-hybrid and biochemical pull-down assays13-16. All these assays determine the association under different physical conditions and their results can complement each other. Moreover, the advantage of this fluorescence assay over other assays is that it can show the regulation of subcellular localization of the cargos by KIF5B (Figures 1 and 2).
The protocol is not limited to KIF5B and can be used to identify cargos of other motor proteins, such as some other kinesin motors3 and some of the myosin motors23,24 used in intracellular transport of cargos3. These motor proteins also contain motor domains for movement along microtubules for kinesin motors and microfilaments for myosin motors, and coiled-coil segments for oligomerization. Most of them form homodimers3,23. Therefore, a similar strategy can be applied to them by creating their motorless dominant negative mutants to identify their cargos.
Disclosures
Free Access publication and production of this article is paid for by Roche.
Acknowledgements
Roche's Publication Grant covered the Free Access publication and production of this article. The author also thanks E. Premkumar Reddy, Richard V. Mettus, Stephen C. Cosenza, Sau Ying Yip and Sol D. Gloria for their support and critical reading of the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| absolute ethanol (200 proof) | Fisher Scientific | BP2818 | |
| DAPI | Sigma-Aldrich | D9542 | |
| Hoechst 33342 | Sigma-Aldrich | B2261 | |
| Opti-MEM-I (transfection medium) | Life Technologies | 51985 | |
| ProLong Diamond Antifade Mountant | Life Technologies | P36961 | |
| formaldehyde, para | Fisher Scientific | O4042-500 | |
| Triton-X100 | Fisher Scientific | BP151-500 | |
| X-tremeGENE 9 DNA Transfection Reagent | Roche | 6365787001 | |
| Taq DNA polymerase | Life Technologies | 10342020 | |
| PCR Grade Nucleotide Mix (dNTP Mix) | Roche | 12111424 | |
| Microscope cover glass | Fisher Scientific | 12-541A | |
| GeneRuler 1 kb Plus DNA ladder | Life Technologies | SM1333 | |
| PureLink Quick Gel Extraction kit | Life Technologies | K210012 | |
| BamHI | New England Biolab | R0136 | |
| SalI | New England Biolab | R0138 | |
| T4 DNA ligase | New England Biolab | M0202T | |
| Ethidium bromide | Thermo Scientific | 17898 | |
| DMEM | Life Technologies | 11995-065 | |
| c-MYC rabbit antibody | Cell Signaling | 5606 | |
| p53 mouse antibody | Santa Cruz | sc-126 | |
| Alexa Fluor 488-conjugated anti-rabbit IgG antibody | Life Technologies | A11008 | |
| Alexa Fluor 488-conjugated anti-mouse IgG antibody | Life Technologies | A11059 | |
| fluorescent microscope | Olympus IX71_Fluoview | ||
| computer software for imaging | cellSens |
References
- Hirokawa, N., Niwa, S., Tanaka, Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 68, 610-638 (2010).
- Yu, Y., Feng, Y. M. The role of kinesin family proteins in tumorigenesis and progression: potential biomarkers and molecular targets for cancer therapy. Cancer. 116, 5150-5160 (2010).
- Verhey, K. J., Hammond, J. W. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 10, 765-777 (2009).
- Hirokawa, N., Noda, Y., Tanaka, Y., Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 10, 682-696 (2009).
- Lee, C. M. Transport of c-MYC by Kinesin-1 for proteasomal degradation in the cytoplasm. Biochim Biophys Acta. 1843, 2027-2036 (2014).
- Bittins, C. M., Eichler, T. W., Hammer, J. A., Gerdes, H. H. Dominant-negative myosin Va impairs retrograde but not anterograde axonal transport of large dense core vesicles. Cell Mol Neurobiol. 30, 369-379 (2010).
- Kimura, T., Watanabe, H., Iwamatsu, A., Kaibuchi, K. Tubulin and CRMP-2 complex is transported via Kinesin-1. J Neurochem. 93, 1371-1382 (2005).
- Lindsay, A. J., McCaffrey, M. W. Myosin Va is required for the transport of fragile X mental retardation protein (FMRP) granules. Biol Cell. 106, 57-71 (2014).
- Rivera, J., Chu, P. J., Lewis, T. L., Arnold, D. B. The role of Kif5B in axonal localization of Kv1 K(+) channels. Eur J Neurosci. 25, 136-146 (2007).
- Roland, J. T., Kenworthy, A. K., Peranen, J., Caplan, S., Goldenring, J. R. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 18, 2828-2837 (2007).
- Uchida, A., Alami, N. H., Brown, A. Tight functional coupling of kinesin-1A and dynein motors in the bidirectional transport of neurofilaments. Mol Biol Cell. 20, 4997-5006 (2009).
- Zadeh, A. D., et al. Kif5b is an essential forward trafficking motor for the Kv1.5 cardiac potassium channel. J Physiol. 587, 4565-4574 (2009).
- Su, Y. Y., et al. KIF5B promotes the forward transport and axonal function of the voltage-gated sodium channel Nav1.8. J Neurosci. 33, 17884-17896 (2013).
- Lalioti, V. S., Vergarajauregui, S., Tsuchiya, Y., Hernandez-Tiedra, S., Sandoval, I. V. Daxx functions as a scaffold of a protein assembly constituted by GLUT4, JNK1 and KIF5B. . J Cell Physiol. 218, 416-426 (2009).
- Cho, K. I., et al. Association of the kinesin-binding domain of RanBP2 to KIF5B and KIF5C determines mitochondria localization and function. Traffic. 8, 1722-1735 (2007).
- Diefenbach, R. J., Diefenbach, E., Douglas, M. W., Cunningham, A. L. The ribosome receptor, p180, interacts with kinesin heavy chain, KIF5B. Biochem Biophys Res Commun. 319, 987-992 (2004).
- Shaner, N. C., et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 22, 1567-1572 (2004).
- Waters, J. C. Live-cell fluorescence imaging. Methods Cell Biol. 114, 125-150 (2013).
- Sanderson, M. J., Smith, I., Parker, I., Bootman, M. D. Fluorescence microscopy. Cold Spring Harb Protoc. 2014, (2014).
- Hopkins, S. C., Vale, R. D., Kuntz, I. D. Inhibitors of kinesin activity from structure-based computer screening. Biochemistry. 39, 2805-2814 (2000).
- Zhang, Y., Xiong, Y. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12, 175-186 (2001).
- Michael, D., Oren, M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 13, 49-58 (2003).
- Kneussel, M., Wagner, W. Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nat Rev Neurosci. 14, 233-247 (2013).
- Maravillas-Montero, J. L., Santos-Argumedo, L. The myosin family: unconventional roles of actin-dependent molecular motors in immune cells. J Leukoc Biol. 91, 35-46 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved