A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Visualizing and Tracking Endogenous mRNAs in Live Drosophila melanogaster Egg Chambers
In This Article
Summary
Here, we present a protocol for the visualization, detection, analysis and tracking of endogenous mRNA trafficking in live Drosophila melanogaster egg chamber using molecular beacons, spinning disc confocal microscopy, and open-source analysis software.
Abstract
Fluorescence-based imaging techniques, in combination with developments in light microscopy, have revolutionized how cell biologists conduct live cell imaging studies. Methods for detecting RNAs have expanded greatly since seminal studies linked site-specific mRNA localization to gene expression regulation. Dynamic mRNA processes can now be visualized via approaches that detect mRNAs, coupled with microscopy set-ups that are fast enough to capture the dynamic range of molecular behavior. The molecular beacon technology is a hybridization-based approach capable of direct detection of endogenous transcripts in living cells. Molecular beacons are hairpin-shaped, internally quenched, single-nucleotide discriminating nucleic acid probes, which fluoresce only upon hybridization to a unique target sequence. When coupled with advanced fluorescence microscopy and high-resolution imaging, they enable one to perform spatial and temporal tracking of intracellular movement of mRNAs. Although this technology is the only method capable of detecting endogenous transcripts, cell biologists have not yet fully embraced this technology due to difficulties in designing such probes for live cell imaging. A new software application, PinMol, allows for enhanced and rapid design of probes best suited to efficiently hybridize to mRNA target regions within a living cell. In addition, high-resolution, real-time image acquisition and current, open source image analysis software allow for a refined data output, leading to a finer evaluation of the complexity underlying the dynamic processes involved in the mRNA's life cycle.
Here we present a comprehensive protocol for designing and delivering molecular beacons into Drosophila melanogaster egg chambers. Direct and highly specific detection and visualization of endogenous maternal mRNAs is performed via spinning disc confocal microscopy. Imaging data is processed and analyzed using object detection and tracking in Icy software to obtain details about the dynamic movement of mRNAs, which are transported and localized to specialized regions within the oocyte.
Introduction
Cell biology studies that visualize dynamic events with spatial and temporal resolution have been made possible by the development of fluorescence-based live cell imaging techniques. Presently, in vivo mRNA visualization is achieved via technologies that are based on RNA aptamer-protein interactions, RNA aptamer-induced fluorescence of organic dyes and nucleic acid probe annealing1,2,3. They all offer high specificity, sensitivity and signal-to-background ratio. However, RNA aptamer-centered approaches require extensive genetic manipulation, where a transgene is engineered to express an RNA with artificial structural motifs that are required for protein or organic dye binding. For example, the MS2/MCP system requires the co-expression of a transgene expressing an RNA construct containing multiple tandem repeats of the binding sequence for the bacteriophage MS2 coat protein (MCP), and another transgene encoding a fluorescent protein fused to MCP4,5. The addition of such secondary structural motifs to the RNA, along with a bulky fluorescently tagged protein, has raised concerns that native RNA processes may be affected6. A technology that addresses this concern and offers additional unique advantages is the nucleic acid-based approach, molecular beacons (MBs). MBs allow for the multiplex detection of endogenous mRNAs, discrimination of single nucleotide variations, and fast kinetics of hybridization with target mRNA7,8. MBs are oligonucleotide probes that remain in a quenched hairpin fold prior to undergoing a fluorogenic conformational change once they hybridize to their targets (Figure 1C)9. Several groups have had success in using MBs to detect both non-coding RNAs (microRNAs and lncRNAs)10,11,12,13, RNA retroviruses14 and dynamic DNA-protein interactions15. They have been successfully employed for imaging in various organisms and tissues, such as zebrafish embryos16, neurons13, tumor tissue17, differentiating cardiomyocytes18, and Salmonella19.
Here we describe the design, delivery and detection approach for endogenous mRNAs in living D. melanogaster egg chambers coupled with a microscopy set-up that is fast enough to capture the dynamic range of active molecular transport. The D. melanogaster egg chamber has served as an ideal multicellular model system for a wide range of developmental studies, from early germline stem cell division and maternal gene expression to the generation of segmental body plan20,21. Egg chambers are easily isolated, large and translucent, and able to withstand hours of ex vivo analysis, making them highly amenable to imaging experiments. Much work has focused on the asymmetric localization of maternal transcripts to discrete subcellular regions prior to being actively translated. In particular, oskar mRNA localization and its subsequent translation at the oocyte's posterior pole must occur in a tightly regulated manner to avoid a lethal bicaudal embryo phenotype22. oskar mRNA is transcribed in the 15 germline cells, called nurse cells, and actively transported through cytoplasmic bridges, called ring canals, into the oocyte, the germline cell that becomes the mature egg and is ultimately fertilized (Figure 1A). The considerable amount of information already available regarding the dynamic recruitment and exchange of protein factors to and from oskar mRNP, along with its long-range intracellular travel, make oskar a preferred candidate to study the many processes of the mRNA life cycle. MBs have been instrumental in revealing details about the process of mRNA localization and deciphering the regulation and function of protein factors that control mRNA transport during Drosophila oogenesis. In particular, by microinjecting MBs into nurse cells and performing live cell imaging experiments, the tracking of endogenous mRNAs is possible8,23.
The roadmap presented here offers the steps of a complete process, from carrying out a live cell imaging experiment using MBs, acquiring imaging data, to performing data analysis to track endogenous mRNA in its native cellular environment. The steps can be modified and further optimized to meet the needs of researchers working with other tissues/cell types within their own lab setting.
Protocol
1. Design of MBs for Live Cell Imaging
- Fold the target RNA sequence to predict the mRNA target’s secondary structure using the “RNA form” from the mfold server (http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form).
- Paste/upload the target sequence in FASTA format, select 5 or 10% sub-optimality (structures with a free energy of folding within 5 or 10% of the MFE value, respectively), and adjust the maximum number of computed foldings accordingly (e.g. larger for 10% sub-optimality).
Note: Inclusion of sub-optimal secondary structures when designing MBs allows for the identification of regions within the target mRNA that may be more flexible or more rigid than as predicted for the minimum free energy (MFE) structure alone, which improves the overall design of MBs suited for live cell imaging. - Select an “immediate job” for mRNA targets of 800 nucleotides (nt), or a “batch job” for mRNA lengths between 801 and 8,000 nt. Save the “ss-count” file as simple text file.
- Paste/upload the target sequence in FASTA format, select 5 or 10% sub-optimality (structures with a free energy of folding within 5 or 10% of the MFE value, respectively), and adjust the maximum number of computed foldings accordingly (e.g. larger for 10% sub-optimality).
- Use the “ss-count” file obtained in step 1.1 as input for the PinMol program (https://bratulab.wordpress.com/software/) with the desired parameters, to design several MBs for the mRNA target (see tutorials describing usage of PinMol program24 at https://bratulab.wordpress.com/tutorial-pinmol-mac/).
- Determine the specificity of selected MBs by performing BLAST analysis: use “blastn” with the appropriate database (e.g. for oskar mRNA-specific MBs use the “refseq-rna” database and the Drosophila melanogaster organism).
- Identify any tissue-specific expression of mRNA target (e.g. for oskar mRNA Flybase> High-Throughput Expression Data> FlyAtlas Anatomy Microarray or modENCODE Anatomy RNA-Seq; http://flybase.org/reports/FBgn0003015) and compare with any positive BLAST hits. Eliminate probes that show >50% cross-homology with other mRNAs that are also expressed in the tissue/cell of interest.
- Select the fluorophore and quencher pair appropriate for the microscopy set-up available to perform live cell imaging (e.g. Cy5/BHQ2)25.
2. MB Synthesis, Purification, and Characterization
- Use in-house synthesis and purification as previously described7, or services from commercial providers, to synthesize and purify one to five MBs (see above note), using the following labeling scheme: [5’(Fluorophore)-(C3 or C6 linker)-(2’-O-methyl MB sequence)-(Quencher)3’]. Purify MBs using reverse-phase HPLC, in house or using the services of the commercial provider.
NOTE: The phosphoramidites used for automated probe synthesis must have the 2’-O-methyl ribonucleotide modification. One can also use chimeras of alternating locked-nucleic acid (LNA) and 2’-O-methyl modifications to increase the stability of a hybrid between a shorter MB and its target mRNA26. - Synthesize DNA oligonucleotides that match the sequence of the targeted RNA region and thus are complementary to the probe region of MBs, for use in in vitro characterization (see steps 2.3 to 2.5; above note). Maximize hybridization of the MB with the DNA-oligonucleotide target mimic, by including on each end of the DNA target four additional nucleotides, as found in the target mRNA sequence.
Note: A more rigorous characterization of the MB’s efficiency to detect the targeted sequence can be performed using in vitro synthesized RNA targets instead of complementary DNA oligonucleotides8. - Perform thermal denaturation of the MB alone, measure its melting temperature (Tm), and confirm that the MB assumes the desired hairpin shape at physiological temperature. We have observed Tm values between 60 and 90 °C.
- Perform thermal denaturation of the MB in the presence of the DNA oligonucleotide target and measure the MB:DNA target hybrid’s Tm, as previously described7. A Tm between 55 and 60 °C is desired for the MB:DNA hybrid.
- Perform in vitro hybridization reactions with the corresponding DNA oligonucleotide target, and determine the efficiency of MB:DNA hybrid formation at physiological temperature, as previously described7. Fast hybridization kinetics with the DNA target mimic is desired, however MBs that do not show high hybridization efficiency with DNA targets may have a better performance with the target mRNA in vitro and/or in vivo.
3. Dissection and Preparation of Individual Egg Chambers for Microinjection
- Feed newly hatched, mated females for 2-3 days with fresh yeast paste.
- Anesthetize flies on a CO2 pad and, using fine tweezers (Dumont #5), transfer 1-2 females into a drop of Halocarbon oil 700 on a glass cover slip.
- Using a pair of tweezers, orient the fly with the dorsal side up under a stereomicroscope. Dissect the female abdomen by making a small incision at the posterior end and gently squeeze the pair of ovaries into the oil.
- Explant the ovaries onto an oil drop on a new coverslip. Gently hold one ovary with one tweezer while pinching off the youngest stages of the ovariole with the other tweezer. oskar mRNA is actively localized at and after mid-oogenesis (stages > 7), and younger egg chambers ( stages < 7) are more difficult to inject and do not survive as long. Slowly drag on the cover slip (with a downward movement) until individual ovarioles or egg chambers are isolated and aligned vertically. Further separate single egg chambers by displacing the unwanted stages from the ovariole egg chain.
NOTE: Ensure that individually teased egg chambers do not float in the oil, and that they adhere to the cover slip. This is important for both successful microinjection and image acquisition.
4. Microinjection of MBs into the Nurse Cells of Egg Chambers
- Prepare the MB solution, using one molecular beacon (e.g. osk2216Cy5), or a mix of two MBs that target different mRNAs and which are labeled with spectrally distinct fluorophores (e.g. osk2216Cy5 and drongo1111Cy3). Use a concentration of 200-300 ng/µL each MB in HybBuffer (50 mM Tris-HCl - pH 7.5, 1.5 mM MgCl2 and 100 mM NaCl). For a cocktail of four MBs labeled with the same fluorophore that are targeting the same mRNA at 200 ng/µL each in HybBuffer (e.g. osk82, osk1236, osk2216). Spin down the MB solution immediately prior to loading the needle for microinjection.
- Select the objective. A 40x oil objective is recommended for finding an appropriate egg chamber and for performing microinjection.
- Mount the coverslip with dissected egg chamber onto the microscope stage. Bring up the objective in the focus position and identify an egg chamber at a mid-to-late developmental stage, that is properly oriented for microinjection (i.e., with the AàP axis perpendicular to the needle tip to allow for easy injection within a nurse cell proximal to the oocyte).
- Load a needle (commercial or prepared in house27) with ~1 µL MB solution (see step 4.1) and connect it to the microinjector. For microinjections in D. melanogaster egg chambers, orient the needle (see Table of Materials) at an angle <45° to the microscope stage (e.g. 30°) to avoid puncturing several nurse cells.
- Set-up the injector with injection pressure of 500-1,000 hPa and compensation pressure of 100-250 hPa (see Table of Materials).
- Slowly move the stage to bring in the field of view an area of the oil drop void of egg chambers.
- Using the micromanipulator joystick, gently lower the needle into the oil drop and bring its tip into focus towards the periphery of the field of view.
- Perform a ‘clean’ function to remove the air from the tip of needle and to ensure that there is flow from the needle.
- Bring the needle to the home position and focus on the egg chamber to be microinjected, then bring the needle back into focus and position it near the edge of the egg chamber.
- Perform a fine adjustment of the objective’s Z-position such that the membrane separating the follicle cells from nurse cells is in focus.
- Insert the needle into a nurse cell and perform injection for 2-5 s.
- Gently remove the needle and retract it to the home position.
- Change the objective to the desired magnification for image acquisition (60-63x or 100x), focus on the egg chamber, and begin acquisition.
5. Acquisition of Data Using a Spinning Disc Confocal Microscope Setup
NOTE: See Table of Materials for our specific setup.
- Set up acquisition protocol to record an XYZCt stack of 8-16-bit images (XYZ = volume, C = channel, t = time).
- Select laser lines for the desired channels (e.g. 641 nm laser for Cy5 and 491 nm for GFP) and acquire the channels sequentially: first the fluorescence signal in each channel and then change the Z position, to allow for proper colocalization analysis.
- Select the Z step (e.g. 0.3 µm), and the top and bottom Z limits (e.g. -2 µm to 2 µm).
- Input the acquisition time and sampling rate (e.g. every 15-30 s for up to 1 h).
- Initiate acquisition.
6. Processing, Data Analysis to Obtain Tracking and Colocalization Information, and Preparation of Video Files
- Image Processing
- Download, unpack, and open Icy, an open community platform for bioimage informatics (http://icy.bioimageanalysis.org/)
- Open the XYZCt stack acquired in step 5: Image/Sequence >File>Open.
- Convert stack to ImageJ: ImageJ> Tools> Convert to IJ, have Detached Mode ON.
- Make a substack (a selection of a range of Z steps and time points to be further analyzed): ImageJ>Image>Stacks>Tools>Make Substack…; select the desired channels, Z-steps and time points.
- Save substack as TIFF file: ImageJ>File>Save As>Tiff…; use this file for subsequent steps.
- Split channels: ImageJ>Image>Color>Split Channels.
- Subtract background either using a background stack: ImageJ>Process>Image Calculator…, or using the Rolling ball option: ImageJ>Process>Subtract Background…, select the Rolling ball radius. Preview the image for the radius selected before selecting “Accept”.
Note: Background signal will mainly arise from improper quenching of the flurorophore. The signal:background ratio (S:B) is often used as an indicator for an MB’s “brightness”, and it is measured from in vitro hybridization experiments of the MB and DNA target oligonucleotide. For example, MBs osk1236 and osk2216 have an S:B of ~81 and ~120, respectively. - Adjust the brightness and contrast for each channel: ImageJ>Image>Adjust>Brightness/Contrast, select Apply.
- Save each channel as a separate TIFF file: ImageJ>File>Save As>Tiff....
- Merge the two channels: ImageJ>Image>Color>Merge Channels…; select the channels. Save the new stack as a new TIFF file (see step 6.1.8).
- Spot Detection and Tracking
- Convert back to Icy: ImageJ>Tools>Convert to Icy.
- A scale bar is automatically overlaid onto the stack upon conversion to Icy, if the scale bar plugin is installed [Search using Plugins>Setup>Online plugin]. If needed, edit the scale bar via Inspector window (right side of screen)>Layer tab>Name>Scale bar.
- Deselect/inactivate the ‘eye’ icon for Scale bar from the Layer tab>Name to remove the scale bar from the original stack. It can be reactivated on the final stack.
- Save the newly processed stack by taking a screenshot using the “camera” icon from the Image Window’s menu bar, “Take a screenshot of current view” and File>Save as>Tiff….
- Determine spot sensitivity, if spot sensitivity parameters have already been determined move onto step 6.2.7.
- Detect spots: select the window with the image or stack to be analyzed, Detection&Tracking>Detection>Spot Detector, and fill in the Settings parameters:
- For Input, select “currentSequenceInputDetection” (default).
- For Pre Processing, select “Channel 0” (default), or desired channel by cross-referencing the number in the Inspector window>Sequence tab.
- For Detector, select “Detect bright spot over dark background;” use “Force use of 2D Wavelets for 3D” only if there are not enough Z-slices in the stacks to perform the analysis. Select “Scale(s)” and “Sensitivity” for each scale (add more scales for larger spots). The Scale and Sensitivity (the larger the number the more sensitive is the detection, a maximum of 140 is suggested by Icy) are trial and error variables, that must be visually checked afterwards and decided upon.
- For Region of Interest, use “ROIfromSequence” (default).
- For Filtering, use “NoFiltering” (default), or select “SizeFiltering” to define the “Range of accepted objects (in pixels)”.
- Output: select XLS or XML output setting (select XML format when using 2007 MS Excel or earlier and there are >65,000 spots). If the spot detector results are used for the tracking analysis, also select “Export to SwimmingPool”.
- Repeat detection of spots using various scale/sensitivity values until all or most of the spots are detected. Record all of the final parameters.
- For colocalization analysis, repeat spot detection for the other channel.
- To track spots, select Detection&Tracking>Tracking>Spot Tracking>Run the Spot Detector with parameters from step 6.2.6., or use “Select detection results here” pull-down menu to select an existing dataset (for this, keep Spot Detector window open from step 6.2.5). Press the “Estimate parameters” button and select the desired target motion in the Parameters estimation pop-up window (e.g. “is both diffusive and directed”). Press the “Run tracking” button.
- Repeat spot detection and tracking for other channels when tracking spots of multichannel stacks, following steps 6.2.6 and 6.2.7, beginning with the stack generated from step 6.2.7.
- To visualize tracks, select Detection&Tracking>Tracking>Track Manager – this window opens automatically upon completion of a tracking run. For “Color Track Processor,” select “Enable” and choose the desired representation of color for the tracks. Relevant track processors can be accessed via the “add Track Processor…” pull-down menu (e.g. select “Track Processor Time Clip,” enable the “Track Clipper” window, and choose the desired number of detections to be displayed before and after the current time point.)
- Save tracks information as an XML track file: Detection&Tracking>Tracking>Track Manager>File>Save as….
- Save results by taking a screenshot using the “camera” icon from the Image Window’s menu bar, “Take a screenshot of current view”. Screenshots can be taken with the detected spots and/or the tracks simply by activating/deactivating the corresponding eye icon(s) found in Inspector window>Layer tab>Name>Overlay wrapper.
- Install the TimeStamp Overlay plugin: Plugins>Setup>Online plugin>TimeStamp Overlay>Install.
- Add timestamp: Plugins>TimeStamp Overlay (New). Follow instructions on the pop-up window (lower right corner of screen) for directions on placing and formatting the time stamp. The time interval can be added/changed in the Inspector window>Sequence tab>Sequence Properties>Edit.
- Save results by taking another screenshot. Save image as 1) Tiff format, and 2) as AVI format; for AVI format first convert to RGB rendering (Image/Sequence>Rendering>RGB image).
- Rotate image to desired orientation: Inspector window>Sequence tab>Canvas>Rotation.
- Save rotated image by “Take a screenshot of the current view”. Ensure the “eye” icon for the scale bar is deselected, as it will also rotate with the image.
- Choose and crop ROI: select Region Of Interest>2D ROI>Choose ROI shape and then create/draw ROI on the image; Image/Sequence>Plane (XY)>Fast crop.
- Colocalization analysis
- Prepare a colocalization protocol; several examples are provided on the Icy website (http://icy.bioimageanalysis.org/protocol/List) (see Supplemental Materials).
- Load colocalization protocol: Tools>Scripting>Protocols>Load, and adjust parameters in the interacting blocks (e.g. in the “Wavelet Spot Detecting” block use parameters determined in step 6.2.6.).
- Measure the size of a particle in pixels, determine the colocalization distance and input it into “Colocalizer” block as “Max distance.”
NOTE: The size of the particle in pixels depends on the detection system. To measure size, zoom into a single particle and manually count the pixels that span the width of the signal across. Average the measurements from at least three particles. The maximum distance to be set for colocalization is the size of the particle in pixels (this represents the maximum sum of the radius of two particles touching). - If desired, select one or more ROIs for colocalization analysis: Region Of Interest>2D ROI>Choose ROI shape>Draw ROI on image.
- Crop ROI(s): Image/Sequence>Plane (XY)>Fast crop.
- Perform colocalization: Protocols editor window>Chosen protocol tab>Run. The final block in Protocols editor window will contain overall colocalization percentage based on spot detection, while information at each time point can be found in the Inspector window> Output tab.
- Track colocalized and single particles by following step 6.2.7 (Track spots).
- Save as described in step 6.2.16.
Results
Using PinMol, several MBs can be designed for one mRNA target (Figure 1B-C). After synthesis and purification, the selected MBs are characterized and compared using in vitro analysis.
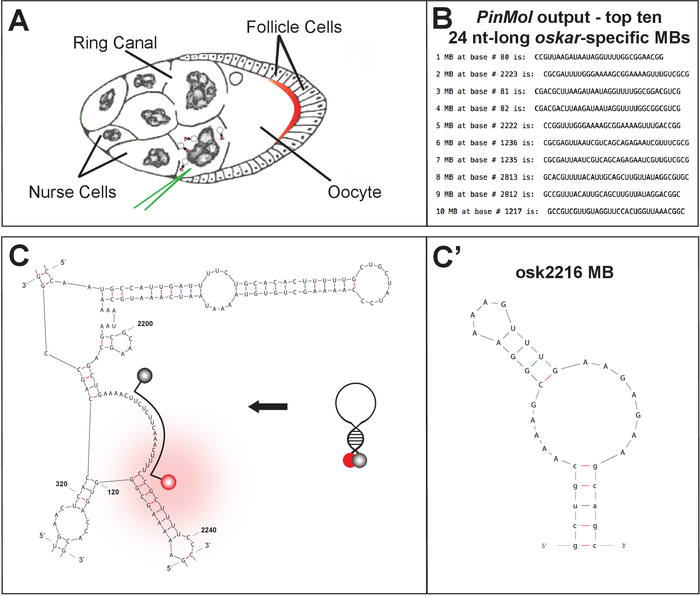
Figure 1: Technique and tissue description for live cell imaging of endogenous mRNAs. ...
Discussion
Live visualization of endogenous mRNA trafficking in Drosophila egg chambers relies on the use of specific, efficient, and nuclease-resistant MBs, which can now be easily designed with PinMol software. MBs are specific probes designed to detect unique sequences within a target mRNA (preferably regions free of secondary structure), making possible highly resolved detection of a transcript. The only limitation when adopting this technique/protocol for other tissues/cell types is the efficiency of MB deliv...
Disclosures
The authors have no conflict of interest to disclose.
Acknowledgements
We thank Salvatore A.E. Marras (Public Health Research Institute Center, Rutgers University) for the synthesis, labeling and purification of molecular beacons, and Daniel St Johnston (The Gurdon Institute, University of Cambridge) for the oskar-MS2/MCP-GFP transgenic fly stock. This work was supported by a National Science Foundation CAREER Award 1149738 and a Professional Staff Congress-CUNY Award to DPB.
Materials
| Name | Company | Catalog Number | Comments |
| Spectrofluorometer | Fluoromax-4 Horiba-Jobin Yvon | n/a | Photon counting spectrofluorometer |
| Quartz cuvette | Fireflysci (former Precision Cells Inc.) | 701MFL | |
| Dumont #5 tweezer | World Precision Instruments | 501985 | Thin tweezers are very important to separate out the individual egg chambers |
| Halocarbon oil 700 | Sigma-Aldrich | H8898 | |
| Cover slip No.1 22 x 40mm | VWR | 48393-048 | |
| Dissecting microscope | Leica MZ6 Leica Microsystems Inc. | n/a | |
| CO2 fruit fly anesthesia pad | Genesee Scienific | 59-114 | |
| Tris-HCL pH7.5 | Sigma-Aldrich | 1185-53-1 | |
| Magnesium chloride | Sigma-Aldrich | 7791-18-6 | |
| NaCl | Sigma-Aldrich | 7647-14-5 | |
| Spinning disc confocal microscope | Leica DMI-4000B inverted microscope equipped with Yokogawa CSU 10 spinning disc Leica Microsystems Inc. | n/a | |
| Hamamatsu C9100-13 ImagEM EMCCD camera | Hamamatsu | n/a | |
| PatchMan NP 2 Micromanipulator | Eppendorf Inc. | 920000037 | |
| FemtoJet Microinjector | Eppendorf Inc. | 920010504 | |
| Injection needle: Femtotips II | Eppendorf Inc. | 930000043 | |
| Loading tip: 20ul Microloader | Eppendorf Inc. | 930001007 | |
| Micro Cover glasses no. 1 or 1.5, 22x40mm | VWR | 48393-026; 48393-172 | |
| Dry yeast | Any grocery store | n/a | |
| Computer, > 20 GB RAM | Although processing can be carried out on most computers, higher capabilities will increase the speed of the processing |
References
- Tyagi, S. Imaging intracellular RNA distribution and dynamics in living cells. Nature Methods. 6 (5), 331-338 (2009).
- Bao, G., Rhee, W. J., Tsourkas, A. Fluorescent probes for live-cell RNA detection. Annual Reviews of Biomedical Engineering. 11, 25-47 (2009).
- Mannack, L. V., Eising, S., Rentmeister, A. Current techniques for visualizing RNA in cells. F1000Research. 5, (2016).
- Larson, D. R., Zenklusen, D., Wu, B., Chao, J. A., Singer, R. H. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 332 (6028), 475-478 (2011).
- Bertrand, E., et al. Localization of ASH1 mRNA particles in living yeast. Molecular Cell. 2 (4), 437-445 (1998).
- Garcia, J. F., Parker, R. MS2 coat proteins bound to yeast mRNAs block 5' to 3' degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. RNA. 21 (8), 1393-1395 (2015).
- Bratu, D. P. Molecular beacons: Fluorescent probes for detection of endogenous mRNAs in living cells. Methods in Molecular Biology. 319, 1-14 (2006).
- Bratu, D. P., Cha, B. J., Mhlanga, M. M., Kramer, F. R., Tyagi, S. Visualizing the distribution and transport of mRNAs in living cells. Proceedings of the National Academy of Sciences of the Unites States of America. 100 (23), 13308-13313 (2003).
- Tyagi, S., Kramer, F. R. Molecular beacons: probes that fluoresce upon hybridization. Nature Biotechnology. 14 (3), 303-308 (1996).
- Chen, M., et al. A molecular beacon-based approach for live-cell imaging of RNA transcripts with minimal target engineering at the single-molecule level. Scientific Reports. 7 (1), 1550 (2017).
- Liu, Y., et al. Multiplex detection of microRNAs by combining molecular beacon probes with T7 exonuclease-assisted cyclic amplification reaction. Analytical and Bioanalytical Chemistry. 409 (1), 107-114 (2017).
- Baker, M. B., Bao, G., Searles, C. D. In vitro quantification of specific microRNA using molecular beacons. Nucleic Acids Research. 40 (2), e13 (2012).
- Ko, H. Y., et al. A color-tunable molecular beacon to sense miRNA-9 expression during neurogenesis. Scientific Reports. 4, 4626 (2014).
- Vet, J. A., et al. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proceedings of the National Academy of Sciences of the Unites States of America. 96 (11), 6394-6399 (1999).
- Li, J., Cao, Z. C., Tang, Z., Wang, K., Tan, W. Molecular beacons for protein-DNA interaction studies. Methods in Molecular Biology. 429, 209-224 (2008).
- Li, W. M., Chan, C. M., Miller, A. L., Lee, C. H. Dual Functional Roles of Molecular Beacon as a MicroRNA Detector and Inhibitor. Journal of Biological Chemistry. 292 (9), 3568-3580 (2017).
- Kuang, T., Chang, L., Peng, X., Hu, X., Gallego-Perez, D. Molecular Beacon Nano-Sensors for Probing Living Cancer Cells. Trends in Biotechnology. 35 (4), 347-359 (2017).
- Ban, K., et al. Non-genetic Purification of Ventricular Cardiomyocytes from Differentiating Embryonic Stem Cells through Molecular Beacons Targeting IRX-4. Stem Cell Reports. 5 (6), 1239-1249 (2015).
- Hadjinicolaou, A. V., Demetriou, V. L., Emmanuel, M. A., Kakoyiannis, C. K., Kostrikis, L. G. Molecular beacon-based real-time PCR detection of primary isolates of Salmonella Typhimurium and Salmonella Enteritidis in environmental and clinical samples. BMC Microbiology. 9, 97 (2009).
- McLaughlin, J. M., Bratu, D. P. Drosophila melanogaster Oogenesis: An Overview. Methods in Molecular Biology. 1328, 1-20 (2015).
- Bastock, R., St Johnston, D. Drosophila oogenesis. Current Biology. 18 (23), R1082-R1087 (2008).
- Rongo, C., Gavis, E. R., Lehmann, R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 121 (9), 2737-2746 (1995).
- Mhlanga, M. M., et al. In vivo colocalisation of oskar mRNA and trans-acting proteins revealed by quantitative imaging of the Drosophila oocyte. PLoS One. 4 (7), e6241 (2009).
- Bayer, L. V., Omar, O. S., Bratu, D. P., Catrina, I. E. PinMol: Python application for designing molecular beacons for live cell imaging of endogenous mRNAs. bioRxiv. , (2018).
- Marras, S. A., Kramer, F. R., Tyagi, S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Research. 30 (21), e122 (2002).
- Bratu, D. P., Catrina, I. E., Marras, S. A. Tiny molecular beacons for in vivo mRNA detection. Methods in Molecular Biology. 714, 141-157 (2011).
- Dean, D. A. Preparation (pulling) of needles for gene delivery by microinjection. Cold Spring Harbor. 2006 (7), (2006).
- Alami, N. H., et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 81 (3), 536-543 (2014).
- Jackson, S. R., et al. Applications of Hairpin DNA-Functionalized Gold Nanoparticles for Imaging mRNA in Living Cells. Methods in Enzymology. 572, 87-103 (2016).
- Zimyanin, V. L., et al. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 134 (5), 843-853 (2008).
- Catrina, I. E., Marras, S. A., Bratu, D. P. Tiny molecular beacons: LNA/2'-O-methyl RNA chimeric probes for imaging dynamic mRNA processes in living cells. ACS Chemical Biology. 7 (9), 1586-1595 (2012).
- Chen, A. K., Behlke, M. A., Tsourkas, A. Efficient cytosolic delivery of molecular beacon conjugates and flow cytometric analysis of target RNA. Nucleic Acids Research. 36 (12), e69 (2008).
- Nitin, N., Santangelo, P. J., Kim, G., Nie, S., Bao, G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Research. 32 (6), e58 (2004).
- Chen, A. K., Behlke, M. A., Tsourkas, A. Avoiding false-positive signals with nuclease-vulnerable molecular beacons in single living cells. Nucleic Acids Research. 35 (16), e105 (2007).
- Bevilacqua, P. C., Ritchey, L. E., Su, Z., Assmann, S. M. Genome-Wide Analysis of RNA Secondary Structure. Annual Review of Genetics. 50, 235-266 (2016).
- Mhlanga, M. M., Vargas, D. Y., Fung, C. W., Kramer, F. R., Tyagi, S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Research. 33 (6), 1902-1912 (2005).
- Eliceiri, K. W., et al. Biological imaging software tools. Nature Methods. 9 (7), 697-710 (2012).
- Bolte, S., Cordelieres, F. P. A guided tour into subcellular colocalization analysis in light microscopy. Journal of Microscopy. 224 (Pt 3), 213-232 (2006).
- Trcek, T., et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nature Commununications. 6, 7962 (2015).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved