Method Article
Focused Ultrasound Induced Blood-Brain Barrier Opening for Targeting Brain Structures and Evaluating Chemogenetic Neuromodulation
In This Article
Summary
This protocol delineates steps necessary for the gene delivery through focused ultrasound blood brain barrier (BBB) opening, evaluation of the resulting gene expression, and measurement of neuromodulation activity of chemogenetic receptors through histological tests.
Abstract
Acoustically Targeted Chemogenetics (ATAC) allows for the noninvasive control of specific neural circuits. ATAC achieves such control through a combination of focused ultrasound (FUS) induced blood-brain barrier opening (FUS-BBBO), gene delivery with adeno-associated viral (AAV) vectors, and activation of cellular signaling with engineered, chemogenetic, protein receptors and their cognate ligands. With ATAC, it is possible to transduce both large and small brain regions with millimeter precision using a single noninvasive ultrasound application. This transduction can later allow for a long-term, noninvasive, device-free neuromodulation in freely moving animals using a drug. Since FUS-BBBO, AAVs, and chemogenetics have been used in multiple animals, ATAC should also be scalable for the use in other animal species. This paper expands upon a previously published protocol and outlines how to optimize the gene delivery with FUS-BBBO to small brain regions with MRI-guidance but without a need for a complicated MRI-compatible FUS device. The protocol, also, describes the design of mouse targeting and restraint components that can be 3D-printed by any lab and can be easily modified for different species or custom equipment. To aid reproducibility, the protocol describes in detail how the microbubbles, AAVs, and venipuncture were used in ATAC development. Finally, an example data is shown to guide the preliminary investigations of studies utilizing ATAC.
Introduction
Use of circuit-specific neuromodulation technologies, such as optogenetics1,2 and chemogenetics3,4,5, has advanced our understanding of psychiatric conditions as neuronal-circuit disorders. Neuronal circuits are difficult to study and even more difficult to control in treating brain disorders because they are typically defined by specific cell types, brain regions, molecular signaling pathways and timing of activation. Ideally for both research and clinical applications, such control would be exerted noninvasively, but achieving both precise and noninvasive neuromodulation is challenging. For example, while neuroactive drugs can reach the brain noninvasively, they lack spatial specificity by acting throughout the brain. On the other hand, electrical deep-brain stimulation can control specific brain regions but has difficulty controlling specific cell types and requires surgery and device placement6.
Acoustically Targeted Chemogenetics7 (ATAC) provides neuromodulation with spatial, cell-type, and temporal specificity. It combines three techniques: focused ultrasound induced blood-brain barrier opening (FUS-BBBO) for spatial targeting, use of adeno-associated viral vectors (AAVs) to noninvasively deliver genes under the control of cell-type specific promoters, and engineered chemogenetic receptors to modulate transfected neural circuits selectively via drug administration. FUS is an FDA-approved technology that takes advantage of ultrasound’s ability to focus deep within tissues, including the human brain, with millimeter spatial precision. At high power, FUS is used for noninvasive targeted ablation, including an FDA-approved treatment for essential tremor8. FUS-BBBO combines low-intensity ultrasound with systemically administered microbubbles, which oscillate in blood vessels at the ultrasound focus, resulting in localized, temporary (6-24 h) and reversible opening of the BBB9. This opening allows for the delivery of proteins9,10, small molecules11, and viral vectors7,12,13,14 to the brain without significant tissue damage in rodents10 and non-human primates15. Clinical trials are ongoing for FUS-BBBO16,17, indicating possible therapeutic applications of this technique.
Viral gene delivery using AAV is also rapidly advancing into clinical use for CNS disorders, with recent FDA and EU regulatory approvals as major milestones. Finally, chemogenetic receptors18, such as Designer Receptors Activated Exclusively by Designer Drugs (DREADDs), are widely used by neuroscientists to provide pharmacological control over neuronal excitation in transgenic or transfected animals19,20. DREADDs are G protein-coupled receptors (GPCRs) that have been genetically engineered to respond to synthetic chemogenetic molecules rather than endogenous ligands, such that systemic administration of these ligands increases or reduces the excitability of DREADD-expressing neurons. When these three technologies are combined into ATAC, they can be used for the noninvasive modulation of selected neural circuits with spatial, cell-type, and temporal precision.
Here, we expand and update a previously published protocol for FUS-BBBO11 by including methodology for accurate targeting of brain regions with FUS-BBBO in mice using simple 3D printed targeting equipment. We, also, show an application of FUS-BBBO to ATAC. We show steps necessary for the delivery of AAVs carrying chemogenetic receptors, and evaluation of gene expression and neuromodulation by histology. This technique is particularly applicable for targeting large or multiple brain regions for gene expression or neuromodulation. For example, a wide area of a cortex can be easily transduced with FUS-BBBO and modulated using chemogenetics. However, gene delivery with an alternative technique, intracranial injections, would require large number of invasive injections and craniotomies. FUS-BBBO and its application, ATAC, can be scaled to animals of different sizes, where brain regions are larger and harder to target invasively.
Protocol
All experiments were conducted under a protocol approved by the Institutional Animal Care and Use Committee of the California Institute of Technology, where data were originally obtained by J.O.S.
1. Design and 3D-printing of animal harness and image guidance hardware
- Use the files from the Szablowski lab website at: https://www.szablowskilab.org/downloads for 3D-printing of the components.
- Ensure that the printing material has low susceptibility in MRI but has an MRI-visible support. See the details of materials used in the materials and reagents section.
- Account for the material degradation with multiple uses by testing the material repeatedly and observing the sites of wear that should be reinforced. Ensure the printed walls are at least 2 mm thick.
- Use high precision 3D printers to improve the targeting precision.
- Counter the gravity and other forces to avoid deviation of plastic 3D printed components by supporting the components along their length and increasing thickness of the 3D printed walls if any bending is observed.
- Account for precision in multiple axes including anterior/posterior, medial/lateral, dorsal/ventral, as well as yaw, pitch, and tilt.
- Test the accuracy of targeting by performing FUS-BBBO and recording the deviation from targeted position.
- If using motorized stereotaxic systems, evaluate for the effects of dynamic movement on the material elasticity by recording the FUS-BBBO targeting procedure on video, and correct any deviations by thickening the 3D printed material walls.
2. Ultrasound system description
- Use an ultrasound system with an eight-element annular array transducer (diameter= 25 mm, natural focal point = 20 mm; aperture (F) = 0.8)) and couple housing to the head with degassed ultrasound gel by applying gel to the shaven mouse head.
NOTE: The center frequency of a transducer used in a previous study7 was 1.5 MHz, pulse duration was 10 ms, and pulse repetition frequency was 1 Hz over 120 s. The pressures were calibrated using optical fiber hydrophone and maintained between 0.36-0.45 MPa. Assume 18% acoustic attenuation through the skull21 for 1.5 MHz and parietal bone. The range of conditions appropriate for a safe BBB opening and AAV delivery have been described elsewhere in detail7,14,22.
3. Animal preparation
- Anesthetize one mouse using isoflurane inhalation at 2% with medical-grade air. Check the depth of anesthesia by a touch pinch to confirm lack of response. Then, apply ophthalmic ointment to prevent corneal drying using a single-use sterile q-tip to prevent cross-contamination of the ointment tube.
NOTE: Typical procedure of FUS-BBBO can range between 30 minutes to 2 hours, and anesthesia must be maintained throughout. - After the mouse is anesthetized, wash a clean catheter with heparinized saline (10 U/ml).
NOTE: An appropriate catheter for a 25-35 g mouse has a 30 G needle and PE10 tubing. - Subsequently disinfect mouse tail with 70% ethanol pad. Place the tail-vein catheter in a lateral tail vein and secure it with a tissue glue. Observe a backflow of blood from the tail vein into the catheter to confirm its placement.
- Shave the mouse head and then use depilation cream after the tissue glue has dried in order to reduce the possibility of air bubbles being trapped under ultrasonic gel during insonation.
- Place the mouse in a 3D-printed MRI carriage, mounting the front teeth on a bite bar and head inside a nosecone (Figure 1a).
- Inject up to 10 µL lidocaine subcutaneously at the site contacting the blunted ear bars. Then, secure the blunted bars to the skull and apply safe amounts of pressure, taking care not to apply pressure to the windpipe as it impedes breathing. Observe the breathing for 30 s to confirm that the animal is breathing freely at a rate of 1/s.
- Connect the targeting guide to ear bars, check breathing as in step 3.6 (Figure 1b), and continue monitoring breathing visually throughout the procedure every minute. A breathing rate elevated above 1/s is one of the indications of loss of anesthesia. Continue monitoring the toe-pinch response every 5 minutes when mice are not in the MRI scanner or if the breathing rate is elevated above 1/s.
- Transfer the MRI carriage into an MRI holder and then inside the bore of a magnet.
NOTE: The design of the hardware is optimized for a 72 mm coil inside a 7T MRI. - Acquire an MRI sequence to localize the mouse in the scanner.
- Select the 3D fast low angle shot (FLASH) sequence to acquire entirety of the brain, using the following parameters, according to specific instructions of the instrument manufacturer. Echo time: 3.9 ms, Repetition time: 15 ms, Excitation pulse angle: 15°, Matrix size: 130 x 130 x 114, Resolution: 350 x 200 x 200 µm per voxel, Averages: 1, Acquisition time: 3 min 42s
- Transfer files from the MRI system onto a computer controlling FUS system.
- Open the imaging sequence in the software to perform MRI-guided targeting, where the image should appear as in Figure 1c.
4. MRI-guided targeting
NOTE: With the use of custom-designed targeting guides, it is not necessary to place ultrasound transducer within an MRI, nor it is necessary to incise the skin to perform targeting by zeroing stereotax on bregma and lambda lines. Follow the steps below to perform the targeting process.
- Place the carriage within a stereotaxic instrument. Secure it in place using a metal block with a double-sided tape and by pressing the carriage against two support posts of the stereotaxic instrument.
- Transfer MRI images to a computer with a running FUS guidance software by selecting files in data manager, right-clicking to bring menu options, and selecting ‘Transfer immediately’.
- Open the FUS guidance software and load image by clicking ‘Open sequence’ and loading all files of the imaging sequence.
- Reformat the image to three axes by pressing right-click and ‘Reformat’.
- Localize the transducer to the circular targeting guide (Figure 1c) by right-clicking.
- In sagittal view, adjust the vertical position of a virtual transducer to account for the thickness of water bath and the transducer housing (in this case – 8.2 mm upwards, Figure 1d).
- Point the area(s) to be targeted in trajectory planner and note the coordinates in a spreadsheet (in this case – midbrain, as in Figure 1d).
- Dial in the desired depth of targeting (z-value in electronic trajectory (Figure 2) and note coordinates in a spreadsheet.
- Target each of the points by pressing ‘Send trajectory’ and ‘Execute’ (Figure 2).
NOTE: This is done when a mouse carrier is placed inside the motors. However, the same targeting can be achieved with high accuracy on a stereotaxic instrument. - To correlate the coordinates of an MRI with the stereotaxic frame, place the custom-mounted transducer over a targeting guide and translate until each of the three targeting bolts (Figure 3, element A) can go through both the transducer holder (Figure 3, element B) and targeting guide (Figure 3, element C). Ascertain that the bolts are not under tension or tilting.
- Translate the transducer 10.56 mm forward in anterior/posterior direction until it is located in the same place where a center of a targeting guide appears on an MRI.
- Determine the distance from a center of the virtual transducer (Figure 3a, element A) to the targeted region (Figure 3a, element B), and move the transducer to these coordinates using stereotaxic frame.
- Proceed to the preparation of injection solution.
5. Injection solution preparation
NOTE: The microbubble solutions are very sensitive to pressure. Consequently, vigorous mixing or rapid injection through thin needles can collapse the microbubbles and reduce efficacy of BBB opening. Additionally, microbubbles are lighter than water and can float to the top of a tube, catheter, or syringe (Figure 4) e.g., in an automatic injector. It is strongly recommended to resuspend microbubble solution immediately before every injection.
- Withdraw 0.8 mL saline using a syringe into 1.5 mL tube.
- Using a syringe, add 0.1 mL of MRI contrast agent into the same 1.5 mL tube and mix.
- Bring the non-activated microbubble23,24 solution to room temperature.
- Right before the insonation, activate microbubbles for 45 s in a microbubble activation device.
- Slowly (over ~3 s) withdraw 0.1 mL of microbubbles using a 1 mL tuberculin syringe and 21 G needle from the middle depth of a liquid.
- Add 0.08 mL of microbubbles into the solution of contrast agent and saline prepared in step 5.3. Mix by tapping with hand for 15 s.
- With the final concentration of AAVs being 0.5-2 x 1010 viral particles per gram of body weight (VP/g), inject the cargo for delivery (in this case AAV9) through 30 G needle into tail vein catheter, in case of ATAC – AAVs carrying chemogenetic receptors, or a negative control, such as AAVs carrying GFP under the same promoter.
- Mix microbubbles by hand for 15 s again to avoid floatation (Figure 4).
- Immediately afterwards, aspirate 200 µL of the microbubble solution through a syringe without a needle attached. Lack of a needle will reduce the shear forces on microbubbles.
- Invert the syringe and mix by pressing the plunger up and down.
- Attach the 30 G needle and, while still inverted, slowly push out the microbubbles until droplets appear at the end of a needle.
6. Insonation procedure
- Set the parameters for insonation: 10 ms pulse duration, 120 repeats, every s, and 0.30-0.45 MPa pressure at the skull.
- Remove the targeting guide, and apply degassed ultrasound gel to the mouse head, making sure to form no bubbles.
- Lower the transducer and place it directly on flat the ear bar holder, and dial in the coordinates into the stereotaxic instruments (Figure 5).
- Inject the solution of AAV (0.5-2 x 1010 VP/g).
- Mix microbubbles and the MRI contrast agent solution for 15 s and inject in 80 µL per 30 g mouse.
- Immediately apply ultrasound for 120 s by pressing ‘Send’ and ‘Execute’.
- If more than one site is targeted, move the transducer to that site and adjust the depth targeting following numbers in the spreadsheet from steps 4.7-4.9. Then repeat steps 6.5-6.6 for every insonated site.
7. MRI evaluation of BBB opening
NOTE: The MRI evaluation of the BBB opening has been described in detail elsewhere11. The location of BBB opening can be visualized as brighter areas in mice that received an injection of a T1-weighted Gd contrast agent.
- After the ultrasound application, record an MRI sequence as in step 3.10.
- Remove the mouse from the MRI scanner, and place it in a recovery cage to allow recovery from anesthesia. Monitor the mice daily for signs of distress, weight loss, or other humane endpoints. Consult veterinary staff and the institutional IACUC guidelines to proceed with any treatment should unexpected adverse events occur.
8. DREADD stimulation with a chemogenetic ligand
- Choose a chemogenetic receptor. For DREADDs, choose the hM3Dq receptor for neuronal activation via Gq coupled pathways19, the hM4Di receptor for inhibition of neuronal activity through Gi/o coupled pathways20, or the KORD receptor for activation of neurons via Gs coupled pathways using Salvinorin-B ligand25.
- Dissolve clozapine-n-oxide (CNO) in sterile saline at concentration of 1 mg/mL. Store the aliquoted CNO at -20 °C.
- Administer CNO19, or another chemogenetic ligand26,27,28, through intraperitoneal route at concentration between 0.3 – 10 mg/kg.
- If effects of CNO on behavior are to be recorded, begin recording the behavioral activity within 15-45 min after the drug administration to achieve the maximum activation of DREADDs19.
- If analysis of neuronal activation is desired, use mice for histological evaluation after 60-120 min post injection.
- Proceed to euthanasia through cardiac perfusion (see section 9).
9. Histological evaluation of gene expression and chemogenetic activation
NOTE: Once the experimental endpoint (e.g., end of behavioral study, time required for gene expression) is achieved, it is critical to confirm the location and presence of the gene expression.
- After activation with a chemogenetic ligand, perform cardiac perfusion to preserve tissues.
- Anesthetize the mouse with Ketamine (100 mg/kg) /Xylazine (10 mg/kg) mixture in sterile saline through an IP injection. Confirm the anesthesia with a toe pinch and ensure the breathing rate has been lowered to approximately 1/s. Provide thermal support through the heating pad until the confirmation of euthanasia.
- Prepare 10% neutral buffered formalin (NBF) and PBS with 10 units of heparin per mL.
NOTE: Solutions should be at 4 °C. - Pour each buffer into separate 50 mL tubes and connect, and prime a peristaltic pump connected to a 25 G butterfly catheter with PBS/heparin solution.
- Attach the limbs to an absorbent blue pad with tape and ensure that the animal is placed in supine position on the pad. Disinfect the fur to avoid cross-contamination of peripheral organs in case they need to be collected.
- Open the peritoneal cavity by a transverse incision, exposing the diaphragm.
- Open the ribcage through two cuts of surgical scissors along the anterior / posterior axis.
- Expose the heart and place needle in a left chamber (right side of the heart as viewed supine) and place in the butterfly catheter in step 9.1.3.
- Make a small incision in the right ventricle to allow blood outflow.
- Turn on the peristaltic pump to begin flushing out the blood with PBS/heparin.
NOTE: If this step is not performed adequately, the blood will clot during fixation with formalin and prevent appropriate perfusion. - After all blood is flushed out and clear PBS starts coming out of the right ventricle, switch the inlet of a peristaltic pump to an NBF solution and begin perfusion for 25 mL per mouse.
- Extract the brain, place in at least 4 mL of NBF and post-fix for 24 h.
- Section the brain using coronal sections on a vibratome using 50 µm section thickness.
- Place each section into a well of a 24 well plate storing the sections throughout the brain.
- Evaluate the expression of the chemogenetic receptors fused to fluorescent proteins (e.g., mCherry or mCitrine), under a fluorescent microscope to identify the sections showing expression to confirm the location. The expression is likely to be dim.
- Perform immunostaining against the fluorophore using the following protocol:
- Place 3 sections in 0.5 mL of solution containing 10% serum of a host of a secondary antibody and incubate for 30 min.
- Transfer the sections into a solution of a primary antibody at 1:250 – 1:1,000 dilution, using 1:500 as a starting point.
- Incubate the sections with primary antibody overnight at 4 °C in a microplate sealed with a paraffin film.
- Wash the sections with PBS, 3x for 5 min at a time.
- Add 0.5 mL per well of the secondary antibody solution in 10% serum.
- Incubate for 4 h in room temperature.
- Wash the sections with PBS, 3x for 5 min at a time.
- Mount on slides with an aqueous mounting medium containing a nuclear stain (e.g., DAPI).
- Evaluate the localization and spread using a confocal microscope by performing a tile scan of an entire section.
- Evaluate intensity of expression by measuring fluorescence pixel intensity in targeted brain regions and compare against an intracranially injected control.
- Alternatively, evaluate percent of positive neurons by counting cells stained against chemogenetic receptors, as compared to DAPI-positive cells, or cell-specific markers.
- Evaluate the tissue damage by performing hematoxylin staining on 50 µm section and imaging for the loss of cells, accumulation of cell debris, and other signs of gross damage.
- To evaluate the specificity of cell-targeting, perform double immunostaining as described below for the chemogenetic receptor and a cell-specific marker. Then, perform cell-positive counts as in step 9.8.
- Place 3 sections in 0.5 mL of a solution containing 10% serum of a host of a secondary antibody and incubate for 30 min.
- Transfer the sections into a solution of a primary antibody against a fluorescent marker of a chemogenetic receptors at 1:250 – 1:1,000 dilution, using 1:500 as a starting point. Add a second primary antibody from a different host species that is targeted against a cell-specific marker of interest (e.g., CamkIIa).
- Incubate the sections with primary antibody overnight at 4 °C in a microplate sealed with paraffin film.
- Wash the sections with PBS, 3x for 5 min at a time.
- Add 0.5 mL per well of secondary antibody solution in 10% serum of the host-species of both secondary antibodies.
NOTE: Each antibody should have a distinct fluorophore and should be reactive against primary antibodies in step 9.10.2. - Incubate for 4 h in room temperature.
- Wash the sections with PBS, 3x for 5 min at a time.
- Mount on slides and fix with an aqueous mounting medium containing a nuclear stain.
10. Evaluate neuronal activation with immunostaining for c-Fos
- Perform c-Fos staining as in point 9.5 of this protocol using a c-Fos primary antibody and a secondary antibody with a fluorescent tag distinct from the nuclear stain.
- Count the percent of cells that are positive for both c-Fos and nuclear stain in the area targeted by a chemogenetic receptor.
- Analyze the percentage of c-Fos positive nuclei in group of mice expressing chemogenetic receptors and treated with a chemogenetic ligand or vehicle control and in group of wild-type mice that are treated with a chemogenetic ligand or vehicle control.
Results
The first step of performing ATAC protocol is the targeting of the FUS-BBBO to the desired brain regions. For example, following the described protocol, the hippocampus was targeted with FUS-BBBO, and contrast agent and AAV9 carrying DREADDs were injected into the mice, followed by a FLASH 3D MRI sequence that acquires images of the mouse brain. A T1 signal enhancement was achieved at the hippocampal region (Figure 6) and in other parts of the brain (Figure 7). After several weeks, DREADDs were expressed inside the target brain region. While many DREADDs are fused to a fluorescent reporter (e.g. mCherry), the process of perfusion and fixation with formaldehyde was found to drastically reduce the fluorescence of these proteins. Immunostaining against mCherry or the DREADD led to more reliable detection of the expression (Figure 8) based on previous experience. In previous experiments, ~85% of the mice showed expression following FUS-BBBO7. A simple test for sufficient levels of expression of DREADDs is testing their functionality on a cellular level. It can be done, for example, by providing a chemogenetic ligand or a saline control, such as CNO19, deschloroclozapine28, or others29, and waiting 2 hours before a cardiac perfusion and fixation. The brain sections were then co-immunostained for c-Fos protein30, which indicates heightened activity of neurons, and for DREADD. The experiment was considered successful, if the site of the brain targeted with DREADDs showed significantly higher number of neuronal nuclei that are c-Fos positive in the group that received a chemogenetic ligand when compared to the group that received saline7 or compared to a contralateral site that was not subjected to FUS-BBBO. Of note, there is a potential for some of these ligands to activate neurons non-specifically without expression of DREADDs. For example, CNO has been shown to be metabolized into low levels of clozapine in mice, which crosses the BBB and activates DREADDs with high potency27. However, it was also shown to bind to non-specific locations. As in every experiment, it is critical to include all proper controls in chemogenetic studies31. One possible control is administration of the chemogenetic ligand to wild-type mice, without procedures, to exclude effects of the drug alone on the desired behavioral or histological assay. Another control could be inclusion of four groups: DREADD + ligand, DREADD + vehicle, EGFP + ligand, EGFP + vehicle, which will account for any potential effects of both gene delivery with FUS-BBBO, and the chemogenetic ligand.
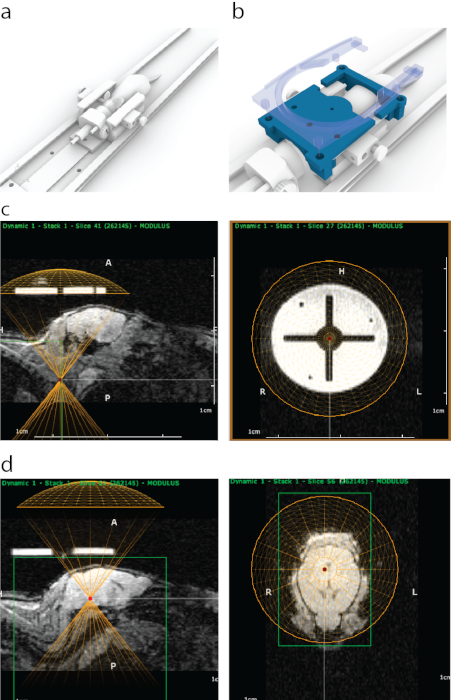
Figure 1: The process of MRI-guided targeting of FUS in ATAC. (a) Mouse placement with ear bars, a nose cone and a platform that can be fit inside an MRI scanner. (b) A 3D-printed guide (blue) that is visible in MRI was attached to ends of ear bar frame and then secured in place with a holder of a surface MRI coil that contains four snap-on bolts (semi-transparent blue). (c) Appearance of the 3D-printed guide in sagittal MRI (left panel), with a bottom of the virtual representation of a transducer aligned (yellow semicircle) with the bottom of the guide. Right panel shows appearance of the 3D-printed guide on MRI from coronal view. The bright circle was made of a polyjet support material that has a strong MRI contrast. The cross was formed with plastic. A yellow circle represents transducer location which was aligned concentrically with the guide inside a stereotaxic frame. (d) To target brain structures a virtual transducer was moved in z-direction above the mice to match the thickness of an ultrasound cone / housing. In this case, because of the thickness of water bath, the transducer was moved 8.2 mm above the guide for accurate targeting. Brain structures were selected using MRI imaging data, and their MRI coordinates were then written down and entered into the stereotaxic machine. Please click here to view a larger version of this figure.
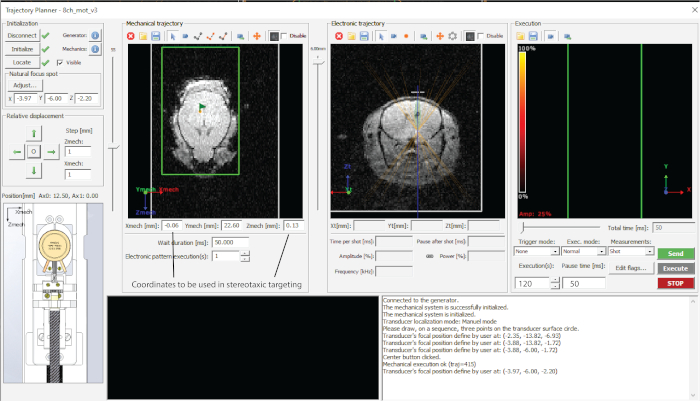
Figure 2: Interface of the software used. Please click here to view a larger version of this figure.
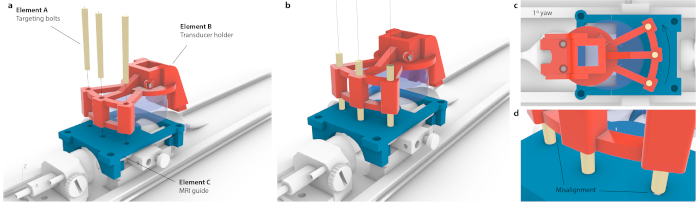
Figure 3: Process of matching MRI coordinate space to stereotaxic instrument. (a) Three holes within a transducer holder were aligned with three holes within the MRI guide, and three conical targeting bolts were inserted without causing flex to the entire assembly. (b) Ideally, all three bolts would sit the center of the holes. (c) If there is any imprecision in alignment, not all three bolts would fit in e.g., in case of small, likely imperceptible yaw of 1°, only one bolt would fit in while the opposite bolts would be stuck at the MRI guide. Alternatively, there could be visible flex of the entire assembly as bolts were forced through. (d) Enlarged view of bolt-fitting. The bolts should be placed concentrically for the best accuracy. Please click here to view a larger version of this figure.
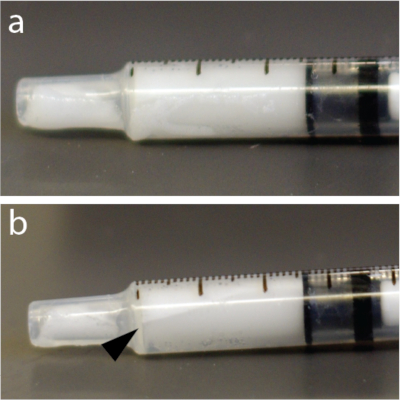
Figure 4: Rapid redistribution of microbubbles within the syringe. (a) Syringe was photographed 5 s after mixing. (b) One minute later, there was a clearly visible layer showing some of the bubbles concentrate near the top of 1 mL tuberculin syringe. This example, in particular, used a solution of microbubbles. Please click here to view a larger version of this figure.
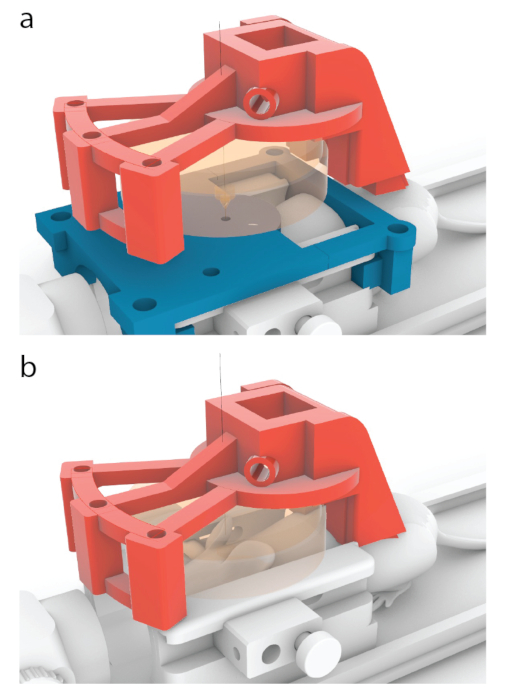
Figure 5: Process of placing the center of a transducer over a center of an MRI guide. (a) In the models shown in this paper, the red carrier has been designed to move 10.56 mm forward from the position shown in Figure 3b, to one shown here. (b) The blue MRI guide was removed before sonication, and an ultrasound gel was applied between the mouse and the transducer (orange) to ensure ultrasound passage. Please click here to view a larger version of this figure.
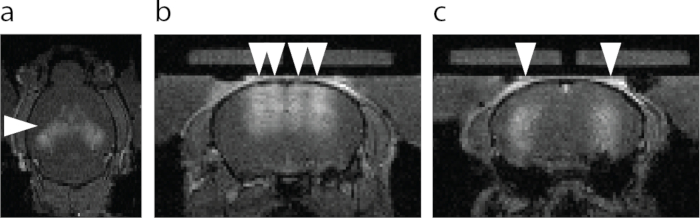
Figure 6: MRI visualization of the BBB opening. (a) Axial view of the BBB opening. Brighter area designated with an arrowhead shows extravasation of an MRI T1 contrast agent. (b) Coronal view of the dorsal hippocampus and the cortex above hippocampus targeted with FUS-BBBO (arrowheads). (c) Coronal view of the central hippocampus targeted with FUS-BBBO (arrowheads). Please click here to view a larger version of this figure.

Figure 7: Example of targeting of 4 brain sites using the three-bolt targeting system described in this paper. Areas with arrowheads showed BBB opened sites with diffusion of an MRI contrast agent. The four sites were targeted in succession, with ~150 s between each BBB opening, from the bottom to top. The image was taken within 2 min after the last BBB opening. Scale bar is 2 mm. Please click here to view a larger version of this figure.
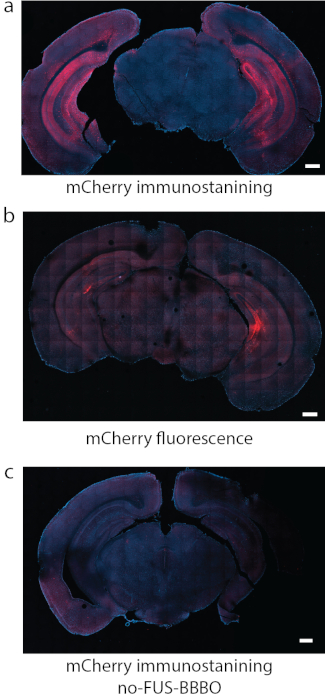
Figure 8: Detection of DREADD expression. (a) Immunostaining for the fluorophore attached to DREADDs, in this case mCherry was a reliable method of detection in some studies. (b) In another representative section with DREADDs targeted to hippocampus using the same conditions as in (a), the fluorescence of mCherry by itself produced strong background and relatively weak signal. (c) As a negative control, a mouse that received systemic injection of AAV, but did not undergo FUS-BBBO, was used. No significant expression can be found by mCherry immunostaining. Scale bars are 500 mm. (Data in a, c adapted from7 with permissions, Copyright 2020 Nature-Springer). Please click here to view a larger version of this figure.
Discussion
ATAC requires successful implementation of several techniques for successful neuromodulation of specific neural circuits, including accurate MRI-guided targeting, FUS-BBBO, and histological evaluation of gene expression. 3D-printable components were developed to simplify targeting of small brain structures with imaging-guided FUS-BBBO.
MRI-guided focused ultrasound (MRIgFUS) administration poses a number of challenges. First, typical MRI coil has limited space that is designed to only accommodate a specimen and not the ultrasound hardware. The larger bores of MRIs increase the cost of equipment and decrease image quality, as the signal is related to the fill factor of a coil32. Consequently, any FUS hardware placed on the top of an animal image in MRI will compromise imaging quality. Second, designing MRI-compatible devices is difficult and expensive. MRI compatible materials need to be diamagnetic, have low propensity of creating eddy currents during radiofrequency irradiation, and have low magnetic susceptibility in high magnetic fields. In any conductive material, the creation of eddy currents or its magnetic susceptibility will also negatively affect imaging quality. Finally, the available MRI-compatible materials have lower Young’s moduli and durability than the metals typically used in production of precise targeting machines e.g., stereotaxic frames. The motors used for positional adjustments need to be MRI-compatible and placed outside of the MRI bore due to their size. These motors have to be connected at a distance to the transducer inside an MRI bore using MRI-compatible materials. Issues of plastic warping, lack of sufficient space inside the bore to implement robustly sized components, and insufficient room for changing targeting positions across the entire brain have affected targeting accuracy in previous work.
To resolve these problems, a decision was made to perform imaging in MRI and FUS-BBBO administration outside of the scanner. To allow for MRI-guidance, mice were placed inside a 3D-printed restraint that had an MRI-visible targeting guide that could be used to localize the mouse brain structures both in the MRI and in the stereotax coordinate space. Since both the mouse skull and the targeting guide are firmly attached to ear bar holders (Figure 1a,b), a targeting guide can be used to correlate spatial coordinates within MRI image and zero the stereotaxic instruments. The restraint does not have moving parts and does not contain a transducer, which allowed us to make it both robust and sufficiently small to fit inside an MRI and removed signal interference from transducer’s electronics. The space inside the targeting guide has been hollowed as the 3D-printed support for some materials is visible in MRI (Figure 1c). Holes in the assembly were introduced to enable stereotax calibration (Figure 3). The ultrasound transducer was attached to an electrode holder of a stereotax, and targeting was performed as described in section 4 (Figure 1d). The transducer should be supported along its length by housing of ear bars, preventing any deviation from the level plane. The targeting in the dorso-ventral direction can be achieved using phase-shifts in an annular array.
The practical targeting precision is determined by ultrasound focusing and skull attenuation. FUS-BBBO procedure has been described in detail for rats11 and has been implemented in a number of other model organisms23,33,34 and in humans16,17. The relationship between ultrasound focus size inversely proportional to frequency, where higher frequencies can result in more precise delivery. However, the attenuation of the skull increases with frequencies35 which may lead to skull heating and damage to the cortical areas. The exact targeting strategy will depend on the brain site. The sites where a full-width half maximum pressure fits within the brain tissue allow for predictable and safe BBB opening in many brain structures such as the striatum, midbrain, and hippocampus. Regions near the base of the brain pose a specific challenge in mice. Mouse brain measures approximately 8-10 mm in dorso-ventral direction, which is comparable to the full-width half maximum size of many commercially available transducers. Consequently, targeting at the bottom of the skull can lead to ultrasound reflection from the bones and air present in ear canals, mouth, or windpipe which can lead to unpredictable patterns of high and low pressures36. Some of these pressures can cross an inertial cavitation threshold which has been shown to cause bleeding and tissue damage37. To target regions which are located near the base of the skull, it may be preferable to use intersectional ATAC7, where intersectional genetics38 is used to restrict gene expression to a smaller area then the one targeted with FUS beam. In the published example of intersectional ATAC, a transgenic animal expressing a gene editing enzyme (Cre38) in dopaminergic cells has been targeted with ultrasound in the subsection of the region containing dopaminergic cells. Finally, the cortical regions can be targeted with FUS, but the diffraction and reflection of ultrasound may occur leading to uneven pressure profiles. This protocol does not cover the targeting of cortical regions as it will be highly dependent on the used species; however, some targeting of the cortex above hippocampus7 (e.g., Figure 7) has been observed indicating that at, least in mice, it is possible.
The choice of a chemogenetic activator and dosing will depend on the specific experimental needs. A number of studies, including one of the authors’ studies7, showed no significant non-specific response39,40, while higher doses (e.g., 10 mg/kg) can produce side effects, at least in some cases41. However, as with all behavioral experiments, proper controls31 are essential due to potential off-targeted activity of CNO and its metabolites42. Such controls could include administration of CNO and saline controls to animals expressing DREADDs and administration of CNO to wild-type animals or in some specific cases a comparison of ipsi- and contralateral sites of the brain that respectively do and do not express chemogenetic receptors. Additionally, recent research revealed a number of new DREADD agonists with improved specificity28,29,43. Other chemogenetic receptors5,25,44 can also be used in conjunction with ATAC procedure.
Histological evaluation of gene expression is necessary post-mortem for every animal. A small fraction of animals show poor gene expression following FUS-BBBO7. Additionally, it is necessary to show the spatial accuracy and specificity of gene expression since mis-targeting is possible. Of note, some AAVs may show retrograde or anterograde tracing capability45 and can cause transfection far from the site targeted with ultrasound despite accurate ultrasound targeting. If the expressed chemogenetic receptor is fused to or co-expresses a fluorophore, imaging of the fluorophore in tissue sections may be sufficient to evaluate localization and intensity of expression. However, many fluorescent proteins are damaged by the tissue fixation process, and immunostaining for mCherry protein that is frequently used with DREADDs yielded better signal in previous studies7. Finally, due to the density of neurons in certain parts of the brain (e.g., granular cell layer in hippocampus), using nuclearly-localized fluorophores expressed under IRES, as opposed to fusions, to perform cell-counts may be beneficial since nuclei can be easily segmented and counterstained with nuclear stains, such as DAPI or TO-PRO-3. To evaluate neuromodulation by c-Fos staining, performing nuclear counterstaining and counting c-Fos positive nuclei, rather than any fluorescence signal, is imperative. In some cases, cellular debris can show fluorescence and confound the measurements of positive cells.
Limitations of the drug and gene delivery with FUS-BBBO include lower resolution than delivery with invasive intracranial injections and the need for larger amounts of injected drugs or viral vectors. Additionally, while a direct injection into the brain results in exclusive delivery to an injected site, FUS-BBBO uses an intravenous route resulting in possible delivery to peripheral tissues. Limitations of using chemogenetics for neuromodulation include a slow timescale, which may be inadequate to some behavioral protocols which require rapid changes in intensity of neuromodulation.
Disclosures
No conflict of interest.
Acknowledgements
This research was supported by Brain and Behavior Foundation, NARSAD Young Investigator Award. Several 3D printed components were originally designed by Fabien Rabusseau (Image Guided Therapy, France). Author thanks John Heath (Caltech) and Margaret Swift (Caltech) for technical help with preparing the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| 21-gauge needles (BD) | Fisher Scientific | 14826C | |
| 25-gauge butterfly catheter | Harvard Bioscience | 725966 | |
| 30-gauge needles (BD) | Fisher Scientific | 14826F | |
| Absorbent blue pad | Office Depot | 902406 | |
| Anti-c-Fos antibody | Santa Cruz Biotechnology | SC-253-G | |
| Anti-mCherry antibody | Thermofisher | PA534974 | |
| Bruker Biospec 70/30 | Bruker | custom | includes the RF coils |
| Clozapine-n-oxide | Tocris | 4936 | |
| Custom designed 3D printed mouse harnesses and MRIgFUS targeting components | ImageGuidedTherapy, Szablowski lab | custom | download from szablowskilab.org/downloads |
| Custom MRIgFUS machine | ImageGuidedTherapy | N/A | |
| Definity microbubbles | Lantheus | DE4 | |
| Degassed aquasonic/ultrasound gel | Fisher Scientific | 5067714 | |
| Depilation crème | Nair | n/a | |
| Eight-element annular array transducer | Imasonic Inc. | custom | |
| Ethanol Pads/Alcohol Swabs (70%) (BD) | Office Depot | 599893 | |
| Heparin | Sigma-Aldrich | H3149-25KU | |
| Isoflurane | Patterson Veterinary | 07-893-1389 | |
| Ketamine | Patterson Veterinary | 07-890-8598 | |
| Neutral buffered formalin (10%) | Sigma-Aldrich | HT501128-4L | |
| Optical fiber hydrophone | Precision Acoustics | ||
| PE10 tubing | Fisher Scientific | NC1513314 | |
| Peristaltic pump | |||
| Phosphate-buffered saline (PBS) | Sigma-Aldrich | 524650-1EA | |
| Prohance contrast agent | Bracco | 0270-1111-04 | |
| Saline | Fisher Scientific | NC9054335 | |
| Secondary antibody, Donkey-anti goat | ThermoFisher | A-11055 | |
| Secondary antibody, Donkey-anti rabbit | ThermoFisher | 84546 | |
| Surgical scissors (straight) | Fisher Scientific | 17467480 | |
| ThermoGuide Software | ImageGuidedTherapy | ||
| Tissue glue (Gluture) | Fisher Scientific | NC9855218 | |
| Tuberculin Syringe (1 mL) (BD) | Fisher Scientific | 14823434 | |
| VeroClear 3D printable material | Stratasys | RGD810 | |
| Vialmix microbubble activation device | Lantheus | VMIX | |
| Vibrating microtome | Compresstome | VF-300 | |
| Xylazine | Sigma-Aldrich | X1251-1G |
References
- Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G., Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience. 8, 1263-1268 (2005).
- Zhang, F., Wang, L. -. P., Boyden, E. S., Deisseroth, K. Channelrhodopsin-2 and optical control of excitable cells. Nature Methods. 3, 785-792 (2006).
- Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S., Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proceedings of the National Academy of Sciences. 104, 5163-5168 (2007).
- Lerchner, W., et al. Reversible silencing of neuronal excitability in behaving mice by a genetically targeted, ivermectin-gated Cl- channel. Neuron. 54, 35-49 (2007).
- Magnus, C. J., et al. Chemical and genetic engineering of selective ion channel-ligand interactions. Science. 333, 1292-1296 (2011).
- Deeb, W., et al. Proceedings of the fourth annual deep brain stimulation think tank: a review of emerging issues and technologies. Frontiers in Integrative Neuroscience. 10, 38 (2016).
- Szablowski, J. O., Lee-Gosselin, A., Lue, B., Malounda, D., Shapiro, M. G. Acoustically targeted chemogenetics for the non-invasive control of neural circuits. Nature Biomedical Engineering. 2, 475-484 (2018).
- Elias, W. J., et al. A pilot study of focused ultrasound thalamotomy for essential tremor. New England Journal of Medicine. 369, 640-648 (2013).
- Burgess, A., Hynynen, K. Noninvasive and targeted drug delivery to the brain using focused ultrasound. ACS Chemical Neuroscience. 4, 519-526 (2013).
- Kinoshita, M., McDannold, N., Jolesz, F. A., Hynynen, K. Noninvasive localized delivery of Herceptin to the mouse brain by MRI-guided focused ultrasound-induced blood-brain barrier disruption. Proceedings of the National Academy of Sciences U. S. A. 103, 11719-11723 (2006).
- Samiotaki, G., Acosta, C., Wang, S., Konofagou, E. E. Enhanced delivery and bioactivity of the neurturin neurotrophic factor through focused ultrasound-mediated blood-brain barrier opening in vivo. Journal of Cerebral Blood Flow & Metabolism. 35, 611-622 (2015).
- O'Reilly, M. A., Waspe, A. C., Chopra, R., Hynynen, K. MRI-guided disruption of the blood-brain barrier using transcranial focused ultrasound in a rat model. Journal of Visualized Experiments. (61), e3555 (2012).
- Thévenot, E., et al. Targeted delivery of self-complementary adeno-associated virus serotype 9 to the brain, using magnetic resonance imaging-guided focused ultrasound. Human gene Therapy. 23, 1144-1155 (2012).
- Hsu, P. -. H., et al. Noninvasive and targeted gene delivery into the brain using microbubble-facilitated focused ultrasound. PloS One. 8, 58682 (2013).
- Wang, S., Olumolade, O. O., Sun, T., Samiotaki, G., Konofagou, E. E. Noninvasive, neuron-specific gene therapy can be facilitated by focused ultrasound and recombinant adeno-associated virus. Gene Therapy. 22, 104 (2015).
- Downs, M. E., et al. Long-Term Safety of Repeated Blood-Brain Barrier Opening via Focused Ultrasound with Microbubbles in Non-Human Primates Performing a Cognitive Task. PLoS One. 10, 0125911 (2015).
- Lipsman, N., et al. Blood-brain barrier opening in Alzheimer's disease using MR-guided focused ultrasound. Nature Communications. 9, 2336 (2018).
- Carpentier, A., et al. Clinical trial of blood-brain barrier disruption by pulsed ultrasound. Science Translational Medicine. 8, 343 (2016).
- Sternson, S. M., Roth, B. L. Chemogenetic Tools to Interrogate Brain Functions. Annual Reviews Neurosciences. 37, 387-407 (2014).
- Alexander, G. M., et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 63, 27-39 (2009).
- Zhu, H., et al. Chemogenetic inactivation of ventral hippocampal glutamatergic neurons disrupts consolidation of contextual fear memory. Neuropsychopharmacology. 39, 1880-1892 (2014).
- Choi, J. J., Pernot, M., Small, S. A., Konofagou, E. E. Noninvasive, transcranial and localized opening of the blood-brain barrier using focused ultrasound in mice. Ultrasound in Medicine & Biology. 33, 95-104 (2007).
- Thévenot, E., et al. Targeted delivery of self-complementary adeno-associated virus serotype 9 to the brain, using magnetic resonance imaging-guided focused ultrasound. Human Gene Therapy. 23, 1144-1155 (2012).
- Hynynen, K., McDannold, N., Vykhodtseva, N., Jolesz, F. A. Noninvasive MR imaging-guided focal opening of the blood-brain barrier in rabbits. Radiology. 220, 640-646 (2001).
- Yang, F. -. Y., Fu, W. -. M., Chen, W. -. S., Yeh, W. -. L., Lin, W. -. L. Quantitative evaluation of the use of microbubbles with transcranial focused ultrasound on blood-brain-barrier disruption. Ultrasonics Sonochemistry. 15, 636-643 (2008).
- Vardy, E., et al. A New DREADD Facilitates the Multiplexed Chemogenetic Interrogation of Behavior. Neuron. 86, 936-946 (2015).
- Thompson, K. J., et al. DREADD Agonist 21 Is an Effective Agonist for Muscarinic-Based DREADDs in Vitro and in Vivo. ACS Pharmacology and Translational Sciences. 1, 61-72 (2018).
- Gomez, J. L., et al. Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science. 357, 503-507 (2017).
- Nagai, Y., et al. Deschloroclozapine: a potent and selective chemogenetic actuator enables rapid neuronal and behavioral modulations in mice and monkeys. bioRxiv. , 854513 (2019).
- Thompson, K. J., et al. DREADD Agonist 21 Is an Effective Agonist for Muscarinic-Based DREADDs in Vitro and in Vivo. ACS Pharmacology and Translational Science. 1, 61-72 (2018).
- Bullitt, E. Expression of c-fos-like protein as a marker for neuronal activity following noxious stimulation in the rat. Journal of Comparative Neurology. 296, 517-530 (1990).
- Mahler, S. V., Aston-Jones, G. CNO Evil? Considerations for the use of DREADDs in behavioral neuroscience. Neuropsychopharmacology. 43, 934 (2018).
- Gruber, B., Froeling, M., Leiner, T., Klomp, D. W. J. RF coils: A practical guide for nonphysicists. Journal of Magnetic Resonance Imaging. 48, 590-604 (2018).
- Treat, L. H., McDannold, N., Vykhodtseva, N., Hynynen, K. Transcranial MRI-guided focused ultrasound-induced blood-brain barrier opening in rats. IEEE. 2, 998-1000 (2004).
- Choi, J. J., Pernot, M., Small, S. A., Konofagou, E. E. Feasibility of transcranial, localized drug-delivery in the brain of Alzheimer's-model mice using focused ultrasound. IEEE. 2, 988-991 (2005).
- Cobbold, R. S. . Foundations of Biomedical ultrasound. , (2006).
- Younan, Y., et al. Influence of the pressure field distribution in transcranial ultrasonic neurostimulation. Medical Physics. 40, 082902 (2013).
- McDannold, N., Vykhodtseva, N., Hynynen, K. Targeted disruption of the blood-brain barrier with focused ultrasound: association with cavitation activity. Physics in Medicine and Biology. 51, 793-807 (2006).
- Branda, C. S., Dymecki, S. M. Talking about a Revolution: The Impact of Site-Specific Recombinases on Genetic Analyses in Mice. Developmental Cell. 6, 7-28 (2004).
- Jendryka, M., et al. Pharmacokinetic and pharmacodynamic actions of clozapine-N-oxide, clozapine, and compound 21 in DREADD-based chemogenetics in mice. Scientific Reports. 9, 4522 (2019).
- Manvich, D. F., et al. The DREADD agonist clozapine N-oxide (CNO) is reverse-metabolized to clozapine and produces clozapine-like interoceptive stimulus effects in rats and mice. Scientific Reports. 8, 3840 (2018).
- Martinez, V. K., et al. Off-Target Effects of Clozapine-N-Oxide on the Chemosensory Reflex Are Masked by High Stress Levels. Frontiers in Physiology. 10, 521 (2019).
- Gomez, J. L., et al. Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science. 357, 503-507 (2017).
- Bonaventura, J., et al. High-potency ligands for DREADD imaging and activation in rodents and monkeys. Nature Communications. 10, 1-12 (2019).
- Roth, B. L. DREADDs for Neuroscientists. Neuron. 89, 683-694 (2016).
- Aschauer, D. F., Kreuz, S., Rumpel, S. Analysis of Transduction Efficiency, Tropism and Axonal Transport of AAV Serotypes 1, 2, 5, 6, 8 and 9 in the Mouse Brain. PLoS One. 8, 76310 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved