A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Detection of Cell-Free DNA in Blood Plasma Samples of Cancer Patients
In This Article
Summary
In this paper we present a detailed protocol for non-invasive liquid biopsy technique, including blood collection, plasma and buffy coat separation, cfDNA and germline DNA extraction, quantification of cfDNA or germline DNA, and cfDNA fragment enrichment analysis.
Abstract
Identifying mutations in tumors of cancer patients is a very important step in disease management. These mutations serve as biomarkers for tumor diagnosis as well as for the treatment selection and its response in cancer patients. The current gold standard method for detecting tumor mutations involves a genetic test of tumor DNA by means of tumor biopsies. However, this invasive method is difficult to be performed repeatedly as a follow-up test of the tumor mutational repertoire. Liquid biopsy is a new and emerging technique for detecting tumor mutations as an easy-to-use and non-invasive biopsy approach.
Cancer cells multiply rapidly. In parallel, numerous cancer cells undergo apoptosis. Debris from these cells are released into a patient’s circulatory system, together with finely fragmented DNA pieces, called cell-free DNA (cfDNA) fragments, which carry tumor DNA mutations. Therefore, for identifying cfDNA based biomarkers using liquid biopsy technique, blood samples are collected from the cancer patients, followed by the separation of plasma and buffy coat. Next, plasma is processed for the isolation of cfDNA, and the respective buffy coat is processed for the isolation of a patient's genomic DNA. Both nucleic acid samples are then checked for their quantity and quality; and analyzed for mutations using next-generation sequencing (NGS) techniques.
In this manuscript, we present a detailed protocol for liquid biopsy, including blood collection, plasma, and buffy coat separation, cfDNA and germline DNA extraction, quantification of cfDNA or germline DNA, and cfDNA fragment enrichment analysis.
Introduction
Technological advances have led to the sequencing of hundreds of cancer genomes and transcriptomes1. This has contributed to understanding landscapes of molecular changes across different cancer types2. Further studies on these landscapes have helped characterize the sequential somatic alterations and gene-gene fusions3 that are involved in cancer or tumor progression, by serially disrupting apoptosis pathways4. Therefore, somatic mutations and gene-gene fusions can provide information about tumors by serving as biomarkers in individual patients for a particular tumor type5, identifying existing primary tumors prognosis6, categorizing secondary tumors based on molecular changes7, and identifying druggable tumor targets8. Such information may facilitate in selecting personalized treatment for cancer patients and in determining positive and negative treatment responses9. However, obtaining tumor material for identifying genomic profiling of tumor tissue is an invasive procedure10. Moreover, a tumor biopsy comprises only a small part of a heterogeneous tumor; and may, therefore, not be representative for the molecular profile of the whole tumor11. Serial monitoring and tumor genotyping require a repeated collection of tumor tissues, which, usually, is not feasible due to the invasiveness of tumor biopsy procedure and the safety issues that arise from such procedures12.
The liquid biopsy technique, on the other hand, has gained tremendous attention in precision oncology over the last decade13,14. It is mainly due to the non-invasiveness of this technique, and the possibility of it being repeated at multiple time points, thereby enabling an easy-to-use and safe monitoring technique for the disease courses15,16. Liquid biopsy is based on a phenomenon that tumor cells multiply rapidly, and simultaneously many of them undergo apoptosis and necrosis. This leads to the release of apoptotic cell debris into the patients' blood, together with the DNA fragments that are cut at precise sizes during apoptosis17. The apoptosis of non-cancerous cells also leads to the release of its cellular debris into the blood, however, the apoptosis rate in these cells is relatively much lower than tumor cells18. The rational of the liquid biopsy technique is to capture tumor-associated molecules such as DNA, RNA, proteins, and tumor cells14,19 which circulate continuously in the blood. Various techniques20 can be used for the analysis of these molecules including Next-Generation Sequencing (NGS), digital droplet polymerase chain reaction (ddPCR), real-time PCR, and enzyme-linked immunosorbent assay (ELISA). Liquid biopsy technique enables identifying biomarkers that are characteristics of tumor cells. These biomarker molecules are not just released from specific parts of a tumor, but rather from all parts of the tumor21. Hence, markers identified in liquid biopsy represents the molecular profiling of an entire heterogeneous tumor, in addition to other tumors in the body, thus, having advantages over the tissue biopsy-based technique22.
The cfDNA has a short half-life time in the circulating blood ranging from a few minutes to 1–2 hours23. However, the short half-life time of cfDNA facilitates real-time analyses by evaluating treatment response and dynamic tumor assessments. The tumor-derived cfDNA levels indicate prognostication of tumor stage/size evidenced by several studies, which showed a relationship between cfDNA levels and the survival outcomes24. Moreover, studies have proved that the cfDNA has a better prediction capacity than existing tumor markers25. The prognostication of cfDNA is even more pronounced after cancer treatment, higher levels of cfDNA following treatment correlates well with a reduced rate of survival, and resistance to treatment. Whereas, lower levels of cfDNA following therapy generally corresponds with positive treatment response. Additionally, the cfDNA facilitates early detection of treatment response than the traditional detection methods.
The cfDNA increases the possibility of early detection of cancer-associated mutations: during early-stage disease15, the onset of symptoms26 and before cancer diagnosis up to 2 years27. As cfDNA is released from multiple tumor regions or foci, its analysis provides a comprehensive view of the tumor genome it represents28. Therefore, the cfDNA enables to detect somatic mutations that might have been missed in the tissue samples29. As intra-tumor heterogeneity and subclonal mutations can be detected by deep sequencing of genomic regions spanning thousands of bases, hence the analysis of the cfDNA enables to uncover specific molecular subtypes with distinct genomic signatures13. To obtain a similar level of information through tissue sample many solid biopsies would have been needed.
Furthermore, the cfDNA levels in patients with a localized disease such as colon, ovarian, and lung cancer after a surgical treatment and/or chemotherapy, demonstrated to be a powerful prognostic marker for cancer recurrence and treatment outcomes20. Moreover, in patients with colon, breast, and lung cancer, analyses of cfDNA from the blood could successfully detect the tumor-specific changes, which led to the precise prediction of recurrence several months in advance13. Furthermore, the treatment resistance markers, such as KRAS mutations in patients with CRC receiving anti-EGFR therapy30; VAFs for genes such as PIK3CA, MED1 or EGFR in patients with breast cancer after the treatment with various therapies31; and EGFR T790M resistance mutation in lung cancer patients treated with EGFR-targeted TKIs32 can also be identified by cfDNA analysis.
In summary, the cfDNA analysis can be used to identify precise biomarkers in the field of oncology13,33. In this protocol, blood samples of 3 glioma patients and 3 healthy controls were processed to obtain genomic DNA from WBCs and cfDNA from the plasma. In glioma cancer, mutations in IDH, TERT, ATRX, EGFR, and TP53 serves as a diagnostic as well as prognostic markers that may help in the early diagnosis of glioma tumors, classifying different types of glioma tumors, guiding the accurate treatment for the individual patient and understanding the treatment response34,35. Mutational status of these genes can be identified using blood-derived cfDNA. In this manuscript, we present a detailed protocol of plasma-derived cfDNA that has been used for studying mutational changes in glioma cancer12. Such cfDNA-based liquid biopsy protocol explained in this article can be used for studying mutational changes in many other types of cancers. Moreover, a recent study has shown that cfDNA-based liquid biopsy can detect 50 different types of cancers36.
Blood sample collection, storage, and shipment are crucial steps in this protocol, as uncontrolled temperature during these steps causes lysis of WBCs, leading to the release of genomic DNA from the WBC into the plasma and causing contamination of the cfDNA sample, which affects the rest of the procedure37. Hemolysis due to uncontrolled temperature can impair downstream sample preparation processes of cfDNA, such as the PCR steps38. The serum contains a high proportion of germline cfDNA rather than plasma, although it presents a large background noise for tumor-associated cfDNA39. Therefore, for isolating tumor-associated cfDNA, plasma is a suitable sample39. Blood drawn in an anti-coagulant containing blood collection tube should be centrifuged immediately or within up to two hours, to separate the plasma and to avoid cfDNA contamination. In this protocol, dedicated commercial cfDNA preservation blood collection tubes are used (see Table of Materials), which are an alternative to anticoagulant containing blood collection tubes. These dedicated blood collection tubes preserve cfDNA and cfRNA, and prevents lysis of WBCs for up to 30 days at ambient temperature, and up to 8 days at 37 °C. This facilitates maintaining the appropriate temperature during a blood sample shipment and until the plasma and WBC are separated40.
There are three types of cfDNA extraction methodologies currently available: phase isolation, silicon-membrane based spin column, and magnetic bead-based isolation41. The silicon-membrane based spin column method yielded a high quantity of cfDNA with high integrity compared to other cfDNA extraction methods42.
The quantitative evaluation of DNA is a fundamental requirement in liquid biopsy, there is a need to develop a simple, affordable, and standardized procedure for their easy implementation and wide usage. Three commonly used methods for cfDNA quantification are spectrophotometric, fluorimetric, and qPCR. The fluorimetric method is proved better over the other methods concerning the accuracy, cost, and ease of conducttion43.
The integrity and purity of the cfDNA can be estimated by either agarose electrophoresis or capillary electrophoresis. Agarose electrophoresis neither shows sensitivity at low concentration of cfDNA nor has high resolution to show precise fragment size of cfDNA. On the other hand, capillary electrophoresis has an advantage over the agarose electrophoresis by overcoming the associated challenges and, therefore, widely used by the researchers for cfDNA fragment size analysis. In this protocol, the fragment size distribution of isolated cfDNA was estimated using an automated capillary electrophoresis instrument (see Table of Materials).
Access restricted. Please log in or start a trial to view this content.
Protocol
Prior to blood collection, informed consent from subjects participating in the research is required and must be obtained. The research described in this manuscript was performed in accordance and compliance with the Rabin Medical Center, Israel ethic committee (ethic code: 0039-17-RMC) and the Faculty of Medicine Der Christian-Albrechts-Universität zu Kiel, Germany ethic committee (ethic code: D 405/14).
1. Blood sample collection and storage in cfDNA or cfRNA preservative tubes
- Properly label the preservation tubes
- Collect ~8 mL of blood into the cf-DNA preservation tube (see Table of Materials), using a blood collection set and a holder, as per the standard institutional protocol for venipuncture as described below.
NOTE: The use of a blood collection set may prevent possible backflow of the blood from the tube.- Align the patient with the arm in a downward position.
- Hold the tube upright, with the cap faced upward, while ensuring that the tube contents do not touch the cap or needle tip.
- As blood starts flowing into the tube, release the tourniquet slowly.
- Immediately after the tube is filled with blood (maximum capacity: 8.4 mL of whole blood), gently invert the tube (turn the wrist of the arm that is holding the tube by 180° downward and back) 5 times to stabilize the sample.
NOTE: Inversion ensures the preservative is mixed uniformly with the sample. However, do not shake the contents again, even before plasma preparation. Insufficient mixing of preservatives with the blood sample leads to destabilization of the contents and the formation of micro clots or hemolysis. At this stage, the protocol can be continued immediately for plasma separation or blood-filled tubes can wait for up to 30 days at ambient temperature (15-25 °C), and up to 8 days at 37 °C.
2. Plasma and buffy coat separation and storage
- Centrifuge the blood-filled preservation tube at 425 x g for 20 min at room temperature to separate plasma.
NOTE: Steps 2.2 and 2.3 should be performed in a biosafety cabinet. - Carefully pipette out the upper plasma layer to a fresh tube in 1 mL aliquots, without disturbing the lower layers.
- Carefully transfer the next layer of buffy coat to a fresh tube (the layer appears as a ring above the RBC pellets), while avoiding RBCs in the lower layer.
- Proceed to step 3 with plasma and step 4 with the buffy coat. If needed store the separated contents at -80 °C.
3. Purification of circulating cfDNA from 1 mL of plasma
NOTE: This step is performed with a commercial kit (see Table of Materials). All buffers are provided with the kit.
- Preparation of buffers and reagents
CAUTION: Do not add acidic solutions or bleach directly to the sample preparation waste. Guanidine salts present in Lysis buffer, Binding buffer, and Wash Buffer-1 when combined with bleach or acids can produce highly reactive compounds.- Binding buffer: Mix 300 mL of Binding buffer concentrate with 200 mL of 100% isopropanol to make 500 mL of working Binding buffer. Store at room temperature.
NOTE: Binding buffer allows the optimal binding of the circulating nucleic acids to the silica membrane. 500 mL of the binding buffer is sufficient for processing 276, 138, 92, 69 or 55 samples of 1, 2, 3, 4 or 5 mL of plasma respectively and is stable for 1 year at room temperature. - Wash Buffer-1: Mix 19 mL of Wash Buffer-1 concentrate with 25 mL of 96–100% ethanol to make 44 mL of working Wash Buffer-1. Store at room temperature.
NOTE: Wash Buffer-1 eliminates the contaminants bound to the silica membrane. 44 mL of working Wash Buffer-1 is sufficient for processing 73 samples of 1/2/3/4/5 mL of plasma and is stable for 1 year at room temperature. - Wash Buffer-2: Mix well 13 mL Wash Buffer-2 concentrate with 30 mL of 96–100% ethanol to make 43 mL of working Wash Buffer-2. Store at room temperature.
NOTE: Wash Buffer-2 eliminates the contaminants bound to the silica membrane. 43 mL of working Wash Buffer-2 is sufficient for processing ~56 samples of 1/2/3/4/5 mL of plasma and is stable for 1 year at room temperature. - To a tube containing 310 μg lyophilized carrier RNA, add 1,550 μL of Elution buffer, to prepare a carrier RNA solution of 0.2 μg/μL. After thoroughly dissolving the carrier RNA, divide the solution to suitable aliquots, and store at –30 °C to –15 °C. Do not freeze-thaw these aliquots more than 3 times. To the Lysis buffer, as shown in Table S1, add the reconstituted carrier RNA dissolved in the Elution buffer.
NOTE: Because carrier RNA does not dissolve directly in Lysis buffer, it needs to be dissolved first in an Elution buffer and then in Lysis Buffer. Firstly, silica membrane-nucleic acids binding is enhanced when there are very few target molecules present in the sample. Secondly, the risk of RNA degradation is reduced because of the presence of large amounts of carrier RNA.
- Binding buffer: Mix 300 mL of Binding buffer concentrate with 200 mL of 100% isopropanol to make 500 mL of working Binding buffer. Store at room temperature.
- Before starting the isolation, bring the columns and samples to room temperature and adjust the sample volumes to 1 mL with sterile phosphate-buffered saline (PBS), if needed. Pre heat 2 water baths or heating blocks that contain 50 mL centrifuge tubes and 2 mL collection tubes to 60 °C and 56 °C, respectively.
- To a 50 mL centrifuge tube, add 100 μL of Proteinase K, 1 mL plasma, and 0.8 mL of Lysis buffer containing 1.0 μg of carrier RNA (prepared in step 3.1.4). Close the centrifuge tube with a cap and mix the contents by pulse-vortexing for 30 s, while ensuring a visible vortex in the tube. Thorough mixing of the contents is important for efficient lysis.
NOTE: Immediately after vortexing, proceed to step 3.4, without delay. - Incubate the solution at 60 °C for 30 min.
- Remove the cap, add 1.8 mL of the binding buffer to the tube, and thoroughly mix with pulse vortexing for 15–30 s after placing the cap.
- Incubate the resulting mixture for 5 min on ice and insert the silica membrane column into the vacuum apparatus that is connected to the vacuum pump. Then, firmly insert a 20 mL tube extender into the open column to prevent sample leakage.
- Carefully pour the incubated mixture into the tube extender of the column and switch on the vacuum pump. After all the lysate mixture completely runs through the columns, switch off the vacuum pump, release the pressure to 0 mbar, and remove and discard the tube extender.
NOTE: Avoiding cross-contamination, the tube extender should be discarded carefully, to prevent its spreading over adjacent columns. - Remove the column from the vacuum apparatus, insert into the collection tube, and centrifuge at 11,000 x g for 30 s at room temperature, to remove any residual lysate. Discard the flow-through.
- Add 600 μL of Wash Buffer-1 into the column, centrifuge at 11,000 x g for 1 min at room temperature, discard the flow-through.
- Add 750 μL of Wash Buffer-2 to the column, centrifuge at 11,000 x g for 1 min at room temperature, and discard the flow-through.
- Add 750 μL of ethanol (96–100%) to the column, centrifuge at 11,000 x g for 1 min at room temperature and discard the flow-through.
- Centrifuge the column at 20,000 x g for 3 min, by placing it in a clean 2 mL collection tube.
- Dry the membrane column assembly completely by placing it into a new 2 mL collection tube with the lid open and incubating at 56 °C for 10 min.
- Place the column in a clean 1.5 mL elution tube. To the center of the column membrane, apply 20–150 μL of Elution buffer and incubate at room temperature for 3 min with the lid closed.
NOTE: Ensure that the Elution buffer is equilibrated to room temperature. In case of using elution buffer less than 50 μL, ensure that it is dispensed carefully onto the center of the membrane. This helps with the complete elution of the bound DNA. However, the elution volume is not fixed and can be changed as per the downstream applications. The recovered eluate can be up to 5 μL and certainly less than the elution volume applied to the column. - Centrifuge the recovered eluate in a microcentrifuge at 20,000 x g for 1 min to elute the nucleic acids, and store at -20 °C.
4. Purification of genomic DNA from buffy coat
NOTE: Commercial kit used in this protocol is mentioned in the Table of Materials. Buffers and reagents mentioned in the below protocol i.e., Lysis buffer A, Lysis buffer B, Wash buffer X, Wash Buffer Y, Proteinase Buffer, Elution buffer, and Proteinase K are part of this commercial kit.
- Preparation of the buffers and reagents
CAUTION: Do not add acidic solutions or bleach directly to the sample preparation waste. Guanidine salts present in Lysis buffer B and Wash buffer X when combined with bleach or acids, can produce highly reactive compounds.- Wash Buffer Y: Mix well 12 mL of Wash Buffer Y concentrate with 48 mL ethanol (96–100%) to obtain 60 mL of working Wash Buffer Y. Store at room temperature.
NOTE: 60 mL of working Wash Buffer Y is sufficient for processing 100 buffy coat samples and is stable for 1 year. - Proteinase K: Prepare Proteinase K solution by dissolving 30 mg lyophilized Proteinase K into 1.35 mL of Proteinase Buffer.
NOTE: Total working solution of Proteinase K is sufficient for processing 52 buffy coat samples. Proteinase K working solution can be stored for at least 6 months at -20 °C.
- Wash Buffer Y: Mix well 12 mL of Wash Buffer Y concentrate with 48 mL ethanol (96–100%) to obtain 60 mL of working Wash Buffer Y. Store at room temperature.
- Steps before initiation of the procedure
- Equilibrate the buffy coat to room temperature.
- Set the heat block or water bath at 56 °C.
- Suspend buffy coat in Lysis buffer A to obtain a final volume of 200 μL. Then add 25 μL of Proteinase K solution, and 200 μL of Lysis buffer B. Mix by vortexing and incubate at 70 °C for 10–15 min. Ensure that the samples are completely covered with the lysis solution.
NOTE: For processing series of samples, Proteinase K and Lysis buffer A may be premixed 10–15 minutes before the procedure, but no longer before that, as Proteinase K self-digests in Lysis buffer A without substrate. - Add 210 μL of 96–100% ethanol to the above mixture and vortex vigorously.
NOTE: The addition of ethanol may form a stringy precipitate; however, this will not affect the DNA isolation. Be sure to load the precipitate also on the column, as shown in the following steps. - Load the entire sample onto the silica column placed in a collection tube. Centrifuge for 1 min at 11,000 x g. Place the column in a new collection tube and discard the previous tube along with flow-through.
NOTE: Repeat the centrifugation step if the sample is not drawn completely through the matrix. - Add 500 μL of Wash buffer X, centrifuge for 1 min at 11,000 x g, and discard the flow-through.
- Place the column into the collection tube, add 600 μL of Wash Buffer Y onto the column, centrifuge for 1 min at 11,000 x g, and discard the flow-through.
- Again, place the column into the collection tube, and centrifuge the column for 1 min at 20,000 x g to dry the silica membrane.
- Incubate the column at room temperature for 1 min, placed into a 1.5 mL microcentrifuge tube, and then add 100 μL of Elution buffer. Then, elute the DNA by centrifuging for 1 min at 11,000 x g and store at -20 °C.
5. Quantification of cfDNA and genomic DNA using fluorometer
- Before starting the protocol, perform the following steps.
- Dilute 2 µL of eluted genomic DNA (from step 4.9) in 1:10 proportions with ultrapure nuclease-free water. Due to expected low concentrations, do not dilute cfDNA samples from step 3.15.
- Equilibrate the assay Standard #1 and assay Standard #2 to room temperature.
- Prepare a total of 6 thin-walled clear tubes of 0.5 mL size.
NOTE: The protocol presented is for quantification of 2 cfDNA and 2 genomic DNA samples, therefore, 4 tubes for 4 samples and this assay requires 2 standards. - Label the lids of the tube.
NOTE: Labeling on the side of the tube could interfere with the reading. Additionally, the assay standard tubes are labeled carefully, since calibration of the fluorometer requires that the standards are in the correct order. - Dilute the assay reagent in 1:200 with the Assay buffer to prepare the working solution. For 4 samples and 2 standards, use 6 µL of assay reagent plus 1,194 µL of Assay buffer to make 1,200 µL (200 µL in each tube) of working solution.
NOTE: Do not use a glass container, instead use a clean plastic tube. Each tube must contain approximately 200 µL of the final volume (an assay standard tube must contain 190 µL of the working solution, and the sample tube must contain 180–199 µL of the working solution). Sufficient working solution must be prepared to accommodate all assay standards and samples. - In the working assay standard tubes, add 190 µL of working solution and 10 µL assay standard and mix the solution by vortexing for 2–3 s. Avoid the formation of bubbles within the solution.
- In the sample tubes, add 198 µL working solution and 2 µL of cfDNA or genomic DNA. Mix the solution by vortexing for 2–3 s, and keep it incubated at room temperature for 2 min.
- In the ‘Home’ screen of the fluorometer instrument, press ‘DNA’ and select ‘dsDNA High Sensitivity Assay’, to display the ‘Standards’ screen, then press ‘Yes’ on the fluorometer ‘Standards’ screen to read the standards.
- In the sample chamber, insert the assay Standard #1 tube, close the lid, and press ‘Read’. Remove the tube once the reading is completed (approximately 3 s) and repeat the same step for Standard #2.
- A sample screen is displayed after the completion of the calibration process, then insert a sample tube and repeat step ‘5.8’. The “sample screen” will then display a value that correspond to the concentration of the sample after dilution in the sample tube.
- For each sample repeat step ‘5.9’, until all samples are read.
- Use the following equation to calculate the actual concentration of the sample.
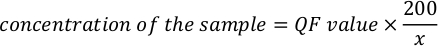
NOTE: The assay values are in ng/mL and corresponds to the concentration after dilution in the assay tube. Equation mentioned in above step 5.11, QF value is the value given by the fluorometer instrument, and x is the number of microliters of sample added to the assay tube. The units for QF values that are generated by the equation are similar to the value provided by the fluorometer. For example, if the value of the fluorometer is in ng/mL, the units for the concentration calculated by the equation are ng/mL.
6. DNA fragment size distribution of cfDNA by fragment analyzer
- The step before initiating the procedure: Equilibrate the DNA dye concentrate and DNA gel matrix to room temperature for 30 min.
- Preparation of the gel-dye mix
CAUTION: Handle solutions with caution as DMSO is known to facilitate the entry of organic molecules into tissues.- Thoroughly thaw the DMSO by vortexing the DNA dye concentrate vial for 10 s. Pipette out 15 µL of this concentrate into a DNA gel matrix vial and store at 4 °C in the dark.
- Again, vortex the capped vial for 10 s until mixing of the gel and dye is visualized.
- Pour the mix on a spin filter to the top receptacle.
- Microcentrifuge the spin filter at 2,240 x g ± 20% for 10 min at room temperature.
- Label the prepared gel-dye in the tube and discard the filter, as per good laboratory practices. Label the tube and record the date of preparation.
NOTE: Discard the filtrate as per good laboratory practices. The gel-dye mix can be used for 5 High Sensitivity (HS) DNA chips. If unused for more than 1 h, store at 4 °C. Storage in the dark is possible for up to 6 weeks.
- To load the gel-dye mix, ensure the position of the base plate of the chip priming station and adjust the clip at the lowest position.
- Equilibrate the gel-dye mix to room temperature for 30 min, while monitoring light exposure.
- Take a new HS DNA chip from a sealed bag and place it on the chip priming station, then remove 9.0 µL of the gel-dye mix and dispense it at the bottom of the chip well, marked as ‘G’.
NOTE: Draw up the gel-dye mix, avoiding particles that may accumulate at the bottom of the vial. While dispensing the gel-dye mix into the HS DNA chip well, insert the tip of the pipette completely, to prevent the formation of large air bubbles. Moreover, touching the pipette at the edges of the well will produce poor results. - Position the plunger at 1 mL and close the chip priming station. Ensure the lock of the latch clicks and set the timer to 60 s, then press the plunger down until it is held by the clip, and exactly after 60 s, release the plunger with the clip-release mechanism.
- When the plunger retreats at least to the 0.3 mL mark, wait for 5 s, and then slowly pull back to the 1 mL position, then open the chip priming station and again remove 9.0 µL of the gel- dye mix and dispense at the bottom of the HS DNA chip well, marked as ‘G’.
- To load the DNA marker, dispense 5 µL of the DNA marker into the well, marked with the ladder symbol. Repeat the procedure for all the 11 sample wells.
- To load the ladder and samples, dispense 1 µL of the DNA ladder in the well, marked with the ladder symbol and then add 1 µL of sample (used wells) or 1 µL of marker (unused wells) in all the 11 sample wells.
- Vortex the HS DNA chip for 60 s at 2,400 rpm by placing the chip horizontally in the adapter. Ensure that the bulge that fixes the HS DNA chip is not damaged during vortexing.
- To insert the HS DNA chip in the fragment analyzer instrument, open the lid and ensure that the electrode cartridge is properly inserted, and the chip selector is positioned to ‘dsHigh Sensitivity DNA’ in the fragment analyzer instrument.
- Carefully mount the HS DNA chip into the receptacle, which fits one way only, then lose the lid by ensuring that the electrode cartridge fits exactly into the wells of the HS DNA chip.
- The display on the fragment analyzer software screen indicates the inserted HS DNA chip and the closed lid, through the chip icon at the top left of the screen.
- To initiate the HS DNA chip run, select the dsDNA High Sensitivity Assay from the ‘Assay’ menu on the instrument screen, then properly fill the table of sample names by feeding information such as sample names and comments and start the chip run by clicking the ‘Start’ button at the upper right of the screen.
- Electrode cleaning after an HS DNA chip run: Immediately remove the used HS DNA chip, as soon as the assay is completed and dispose of it according to good laboratory practices. Perform the following procedure to ensure the electrodes are clean, without leftover residues from the previous assay.
- Fill slowly 350 µL of deionized analysis-grade water into one of the electrode cleaner wells and place the electrode cleaner in the fragment analyzer instrument by opening the lid and then close the lid and wait for about 10 s.
- Remove the electrode cleaner by opening the lid and wait for another 10 s, for the water on the electrodes to evaporate before closing the lid.
Access restricted. Please log in or start a trial to view this content.
Results
Plasma Separation
8.5-9 mL blood collected in cfDNA or cfRNA preservative tubes yields around ~4 mL plasma in volume. The volume of plasma separated from blood collected in EDTA tubes may vary depending on the temperature. Exposure of EDTA tubes containing blood at a temperature higher than 37 °C leads to decreased plasma volume yield44.
Fluorometer Assay Results
cfDNA concentration in 1 mL plasma of each of glioma pati...
Access restricted. Please log in or start a trial to view this content.
Discussion
The collection of a patient’s blood in a tube, shipment and storage are crucial initial steps in liquid biopsy. Improper handling can impair the quality of the plasma and, therefore, can interfere with the results of the liquid biopsy47. If a blood sample is collected in an EDTA blood tube, the plasma must be separated within two hours of blood collection to avoid lysis of WBCs and release of its genomic DNA into the plasma48. WBCs can also undergo apoptosis in an EDT...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgements
The authors would like to thank the members of the Laboratory of Cancer Genomics and Biocomputing of Complex Diseases for their keen observational inputs and their participation in multiple discussions at different stages of this project. The funding support includes Israel Cancer Association (ICA grant for M.F-M 2017-2019) and Kamin grant of Israel Innovation Authority (for M.F-M.).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 2100 Bioanalyzer Instrument | Agilent Technologies, Inc. | G2939BA | The 2100 Bioanalyzer system is an established automated electrophoresis tool for the sample quality control of biomolecules. |
| Adjustable Clip for Priming Station | Agilent Technologies, Inc. | 5042-1398 | Used in combination with syringe to apply defined pressure for chip priming. |
| Agilent High Sensitivity DNA Kit | Agilent Technologies, Inc. | 5067-4626 | The High Sensitivity DNA assays are often used for sample quality control for next-generation sequencing libraries |
| cf-DNA/cf-RNA Preservative Tubes | Norgen Biotek Corp. | 63950 | Norgen's cf-DNA/cf-RNA Preservative Tubes are closed, evacuated plastic tubes for the collection and the preservation of cf-DNA, circulating tumor DNA, cf-RNA and circulating tumor cells in human whole blood samples during storage and shipping |
| Chip Priming Station | Agilent Technologies, Inc. | 5065-4401 | Used to load gel matrix into a chip with a syringe provided with each assay kit— used for RNA, DNA, and protein assays. Includes priming station, stop watch, and 1 syringe clip |
| Electrode Cleaner Kit | Agilent Technologies, Inc. | 5065-9951 | Prevents cross-contamination. Removes bacterial or protein contaminants from electrodes. |
| Filters for Gel Matrix | Agilent Technologies, Inc. | 185-5990 | Used for proper mixing of DNA dye concentrate and DNA gel matrix |
| IKA Basic Chip Vortex | IKA-Werke GmbH & Co. KG | MS-3-S36 | Used for proper mixing of DNA ladder and DNA sample on Bioanalyzer assay chips |
| NucleoSpin Tissue kit | MACHEREY-NAGEL | 740952.5 | With the NucleoSpin Tissue kit, genomic DNA can be prepared from tissue, cells (e.g., bacteria), and many other sources. |
| QIAamp circulating nucleic acid kit | Qiagen | 55114 | The QIAamp Circulating Nucleic Acid Kit enables efficient purification of these circulating nucleic acids from human plasma or serum and other cell-free body fluids. |
| QIAvac 24 Plus vacuum manifold | Qiagen | 19413 | The QIAvac 24 Plus vacuum manifold is designed for vacuum processing of QIAGEN columns in parallel. |
| QIAvac Connecting System | Qiagen | 19419 | In combination with the QIAvac Connecting System, the QIAvac 24 Plus vacuum manifold can be used as a flow-through system. The sample flow-through, containing possibly infectious material, is collected in a separate waste bottle. |
| Qubit 2.0 fluorometer | Invitrogen | Q32866 | The Qubit 2.0 Fluorometer is an easy-to-use, analytical instrument designed to work with the Qubit assays for DNA, RNA, and protein quantitation. |
| Qubit assay tubes | Thermo Fisher Scientific | Q32856 | Qubit assay tubes are 500 µL thin-walled polypropylene tubes for use with the Qubit Fluorometer. |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32851 | The Qubit dsDNA HS (High Sensitivity) Assay Kit is designed specifically for use with the Qubit Fluorometer. The assay is highly selective for double-stranded DNA (dsDNA) over RNA and is designed to be accurate for initial sample concentrations from 10 pg/µL to 100 ng/µL. |
| Vacuum Pump | Qiagen | 84010 | used for vacuum processing of QIAGEN columns |
| Miscellaneous | |||
| 50 ml centrifuge tubes | |||
| Crushed ice | |||
| Ethanol (96–100%) | |||
| Heating block or similar at 56 °C (capable of holding 2 ml collection tubes) | |||
| Isopropanol (100%) | |||
| Microcentrifuge | |||
| Phosphate-buffered saline (PBS) | |||
| Pipettes (adjustable) | |||
| Sterile pipette tips (pipette tips with aerosol barriers are recommended to help prevent cross-contamination) | |||
| Water bath or heating block capable of holding 50 mL centrifuge tubes at 60 °C |
References
- Campbell, P. J., et al. Pan-cancer analysis of whole genomes. Nature. 578 (7793), 82-93 (2020).
- Liotta, L., Petricoin, E. Molecular profiling of human cancer. Nature Reviews Genetics. 1 (1), 48-56 (2000).
- Balamurali, D., et al. ChiTaRS 5.0: the comprehensive database of chimeric transcripts matched with druggable fusions and 3D chromatin maps. Nucleic Acids Research. 48 (1), 825-834 (2019).
- Trédan, O., et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: Analysis from the ProfiLER trial. Annals of Oncology. 30 (5), 757-765 (2019).
- Oliveira, K. C. S., et al. Current perspectives on circulating tumor DNA, precision medicine, and personalized clinical management of cancer. Molecular Cancer Research. 18 (4), 517-528 (2020).
- Siegal, T. Clinical impact of molecular biomarkers in gliomas. Journal of Clinical Neuroscience. 22 (3), 437-444 (2015).
- Komori, T. The 2016 WHO Classification of Tumours of the Central Nervous System: The Major Points of Revision. Neurologia medico-chirurgica. 57 (7), 301-311 (2017).
- Duffy, M. J., O'Donovan, N., Crown, J. Use of molecular markers for predicting therapy response in cancer patients. Cancer Treatment Reviews. 37 (2), 151-159 (2011).
- Saenz-Antoñanzas, A., et al. Liquid Biopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers. 11 (7), 950(2019).
- Marrugo-Ramírez, J., Mir, M., Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. International Journal of Molecular Sciences. 19 (10), 2877(2018).
- Pantel, K., Alix-Panabières, C. Liquid biopsy in 2016: Circulating tumour cells and cell-free DNA in gastrointestinal cancer. Nature Reviews Gastroenterology & Hepatology. 14 (2), 73-74 (2017).
- Bronkhorst, A. J., et al. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomolecular Detection and Quantification. 17, 100087(2019).
- Cescon, D. W., Bratman, S. V., Chan, S. M., Siu, L. L. Circulating tumor DNA and liquid biopsy in oncology. Nature Cancer. 1 (3), 276-290 (2020).
- Palmirotta, R., et al. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Therapeutic Advances in Medical Oncology. 10, 175883591879463(2018).
- Cohen, J. D. J., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 359 (6378), New York, N.Y. 926-930 (2018).
- Wan, J. C. M., et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nature Reviews Cancer. 17 (4), 223-238 (2017).
- Heitzer, E., Haque, I. S., Roberts, C. E. S., Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nature Reviews Genetics. 20 (2), 71-88 (2018).
- Thierry, A. R., et al. Origins, structures, and functions of circulating DNA in oncology. Cancer and Metastasis Reviews. 35 (3), 347-376 (2016).
- Buscail, E., et al. High Clinical Value of Liquid Biopsy to Detect Circulating Tumor Cells and Tumor Exosomes in Pancreatic Ductal Adenocarcinoma Patients Eligible for Up-Front Surgery. Cancers. 11 (11), 1656(2019).
- Heitzer, E., Ulz, P., Geigl, J. B. Circulating Tumor DNA as a Liquid Biopsy for Cancer. Clinical Chemistry. 61 (1), 112-123 (2015).
- Kustanovich, A., Schwartz, R., Peretz, T., Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biology and Therapy. 20 (8), 1057-1067 (2019).
- Crowley, E., Di Nicolantonio, F., Loupakis, F., Bardelli, A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature Reviews Clinical Oncology. 10 (8), 472-484 (2013).
- Celec, P., et al. Cell-free DNA: the role in pathophysiology and as a biomarker in kidney diseases. Expert Reviews in Molecular Medicine. 20, (2018).
- Gautschi, O., et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Letters. 254 (2), 265-273 (2007).
- Bidard, F., et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. International Journal of Cancer. 134 (5), 1207-1213 (2013).
- Chan, K. C. A., et al. Analysis of Plasma Epstein–Barr Virus DNA to Screen for Nasopharyngeal Cancer. New England Journal of Medicine. 377 (6), 513-522 (2017).
- Mao, L., et al. Detection of Oncogene Mutations in Sputum Precedes Diagnosis of Lung Cancer. Cancer Research. 54 (7), 1634-1637 (1994).
- De Mattos-Arruda, L., et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nature communications. 6 (1), 8839(2015).
- Khier, S., Lohan, L. Kinetics of circulating cell-free DNA for biomedical applications: critical appraisal of the literature. Future science OA. 4 (4), 295(2018).
- Misale, S., et al. Resistance to Anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discovery. 4 (11), 1269-1280 (2014).
- Beddowes, E., Sammut, S. J., Gao, M., Caldas, C. Predicting treatment resistance and relapse through circulating DNA. Breast. 34, 31-35 (2017).
- Sacher, A. G., et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA oncology. 2 (8), 1014-1022 (2016).
- Vaidyanathan, R., et al. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab on a Chip. 19 (1), 11-34 (2019).
- Kelly, P. Gliomas: Survival, origin and early detection. Surgical Neurology International. 1 (1), 96(2010).
- Faria, G., Silva, E., Da Fonseca, C., Quirico-Santos, T. Circulating Cell-Free DNA as a Prognostic and Molecular Marker for Patients with Brain Tumors under Perillyl Alcohol-Based Therapy. International Journal of Molecular Sciences. 19 (6), 1610(2018).
- Liu, M. C., et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Annals of Oncology. 31 (6), 745-759 (2020).
- Enko, D., Halwachs-Baumann, G., Kriegshäuser, G. Plasma free DNA: Evaluation of temperature-associated storage effects observed for roche cell-free DNA collection tubes. Biochemia Medica. 29 (1), 153-156 (2019).
- Streleckiene, G., et al. Effects of Quantification Methods Isolation Kits, Plasma Biobanking, and Hemolysis on Cell-Free DNA Analysis in Plasma. Biopreservation and Biobanking. 17 (6), 553-561 (2019).
- Thress, K. S., et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nature Medicine. 21 (6), 560-562 (2015).
- Ward Gahlawat, A., et al. Evaluation of Storage Tubes for Combined Analysis of Circulating Nucleic Acids in Liquid Biopsies. International Journal of Molecular Sciences. 20 (3), 704(2019).
- Lu, J. L., Liang, Z. Y. Circulating free DNA in the era of precision oncology: Pre- and post-analytical concerns. Chronic Diseases and Translational Medicine. 2 (4), 223-230 (2016).
- Iyapparaj, P., et al. Optimization of bacteriocin production by Lactobacillus sp. MSU3IR against shrimp bacterial pathogens. Aquatic Biosystems. 9 (1), 12(2013).
- Ponti, G., et al. The value of fluorimetry (Qubit) and spectrophotometry (NanoDrop) in the quantification of cell-free DNA (cfDNA) in malignant melanoma and prostate cancer patients. Clinica Chimica Acta. 479, 14-19 (2018).
- Medina Diaz, I., et al. Performance of Streck cfDNA blood collection tubes for liquid biopsy testing. PLoS One. 11 (11), 0166354(2016).
- Perkins, G., et al. Multi-Purpose Utility of Circulating Plasma DNA Testing in Patients with Advanced Cancers. PLoS One. 7 (11), 47020(2012).
- Mouliere, F., et al. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Molecular Oncology. 8 (5), 927-941 (2014).
- Trigg, R. M., Martinson, L. J., Parpart-Li, S., Shaw, J. A. Factors that influence quality and yield of circulating-free DNA: A systematic review of the methodology literature. Heliyon. 4 (7), 00699(2018).
- Risberg, B., et al. Effects of Collection and Processing Procedures on Plasma Circulating Cell-Free DNA from Cancer Patients. Journal of Molecular Diagnostics. 20 (6), 883-892 (2018).
- Markus, H., et al. Evaluation of pre-analytical factors affecting plasma DNA analysis. Scientific Reports. 8 (1), 7375(2018).
- Chen, Z., et al. Comprehensive Evaluation of the Factors Affecting Plasma Circulating Cell-Free DNA Levels and Their Application in Diagnosing Nonsmall Cell Lung Cancer. Genetic Testing and Molecular Biomarkers. 23 (4), 270-276 (2019).
- Schwarzenbach, H., Hoon, D. S. B., Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nature Reviews Cancer. 11 (6), 426-437 (2011).
- Malyuchenko, N. V., et al. PARP1 Inhibitors: antitumor drug design. Acta Naturae. 7 (3), 27-37 (2015).
- Fleischhacker, M., Schmidt, B. Circulating nucleic acids (CNAs) and cancer-A survey. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1775 (1), 181-232 (2007).
- Jung, K., Fleischhacker, M., Rabien, A. Cell-free DNA in the blood as a solid tumor biomarker—A critical appraisal of the literature. Clinica Chimica Acta. 411 (21-22), 1611-1624 (2010).
- Frenkel-Morgenstern, M., et al. ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Research. 41, Database issue 142-151 (2013).
- Frenkel-Morgenstern, M., et al. ChiTaRS 2.1--an improved database of the chimeric transcripts and RNA-seq data with novel sense-antisense chimeric RNA transcripts. Nucleic Acids Research. 43, Database issue 68-75 (2015).
- Gorohovski, A., et al. ChiTaRS-3.1-the enhanced chimeric transcripts and RNA-seq database matched with protein-protein interactions. Nucleic Acids Research. 45 (1), 790-795 (2017).
- Tate, J. G., et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Research. 47 (1), 941-947 (2019).
- Brennan, C. W., et al. The somatic genomic landscape of glioblastoma. Cell. 155 (2), 462-477 (2013).
- Szopa, W., Burley, T. A., Kramer-Marek, G., Kaspera, W. Diagnostic and Therapeutic Biomarkers in Glioblastoma: Current Status and Future Perspectives. BioMed Research International. 2017, 1-13 (2017).
- Salesse, S., Verfaillie, C. M. BCR/ABL: from molecular mechanisms of leukemia induction to treatment of chronic myelogenous leukemia. Oncogene. 21 (56), 8547-8559 (2002).
- Frenkel-Morgenstern, M., Valencia, A. Novel domain combinations in proteins encoded by chimeric transcripts. Bioinformatics. 28 (12), 67-74 (2012).
- Frenkel-Morgenstern, M., et al. Chimeras taking shape: Potential functions of proteins encoded by chimeric RNA transcripts. Genome Research. 22 (7), 1231-1242 (2012).
- Simon, M., et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro-Oncology. 17 (1), 45-52 (2015).
- Waitkus, M. S., Diplas, B. H., Yan, H. Isocitrate dehydrogenase mutations in gliomas. Neuro-Oncology. 18 (1), 16-26 (2016).
- Kindler, T., Meyer, R. G., Fischer, T. BCR-ABL as a target for novel therapeutic interventions. Expert Opinion on Therapeutic Targets. 6 (1), 85-101 (2002).
- Overman, M. J., et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed. Journal of Clinical Oncology Official Journal of the American Society of Clinical Oncology. 31 (1), 17-22 (2013).
- Vanderlaan, P. A., et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer. 84 (1), Amsterdam, Netherlands. 39-44 (2014).
- Stewart, C. M., Tsui, D. W. Y. Circulating cell-free DNA for non-invasive cancer management. Cancer Genetics. 228-229, 169-179 (2018).
- Normanno, N., et al. The liquid biopsy in the management of colorectal cancer patients: Current applications and future scenarios. Cancer Treatment Reviews. 70, 1-8 (2018).
- Hufnagl, C., et al. Evaluation of circulating cell-free DNA as a molecular monitoring tool in patients with metastatic cancer. Oncology Letters. 19 (2), 1551-1558 (2020).
- Petit, J., et al. Cell-Free DNA as a Diagnostic Blood-Based Biomarker for Colorectal Cancer: A Systematic Review. The Journal of Surgical Research. 236, 184-197 (2019).
- Poulet, G., Massias, J., Taly, V. Liquid Biopsy: General Concepts. Acta Cytologica. 63 (6), 449-455 (2019).
- Alix-Panabières, C. The future of liquid biopsy. Nature. 579 (7800), 9(2020).
- Eisenstein, M. Could liquid biopsies help deliver better treatment. Nature. 579 (7800), 6-8 (2020).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved