A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Ensemble Force Spectroscopy by Shear Forces
In This Article
Summary
Ensemble force spectroscopy (EFS) is a robust technique for mechanical unfolding and real-time sensing of an ensemble set of biomolecular structures in biophysical and biosensing fields.
Abstract
Single-molecule techniques based on fluorescence and mechanochemical principles provide superior sensitivity in biological sensing. However, due to the lack of high throughput capabilities, the application of these techniques is limited in biophysics. Ensemble force spectroscopy (EFS) has demonstrated high throughput in the investigation of a massive set of molecular structures by converting mechanochemical studies of individual molecules into those of molecular ensembles. In this protocol, the DNA secondary structures (i-motifs) were unfolded in the shear flow between the rotor and stator of a homogenizer tip at shear rates up to 77796/s. The effects of flow rates and molecular sizes on the shear forces experienced by the i-motif were demonstrated. The EFS technique also revealed the binding affinity between DNA i-motifs and ligands. Furthermore, we have demonstrated a click chemistry reaction that can be actuated by shear force (i.e., mechano-click chemistry). These results establish the effectiveness of using shear force to control the conformation of molecular structures.
Introduction
In single-molecule force spectroscopy1 (SMFS), the mechanical properties of individual molecular structures have been studied by sophisticated instruments such as the atomic force microscope, optical tweezers, and magnetic tweezers2,3,4. Restricted by the same directionality requirement of the molecules in the force-generating/detecting setups or the small field of view in magnetic tweezers and the miniature centrifuge force microscope (MCF)5,6,7,8, only a limited number of molecules can be investigated simultaneously using SMFS. The low throughput of SMFS prevents its wide application in the molecular recognition field, which requires the involvement of a large set of molecules.
Shear flow provides a potential solution to apply forces to a massive set of molecules9. In a liquid flow inside a channel, the closer to the channel surface, the slower the flow rate10. Such a flow velocity gradient causes shear stress that is parallel to the boundary surface. When a molecule is placed in this shear flow, the molecule reorients itself so that its long axis aligns with the flow direction, as the shear force is applied to the long axis11. As a result of this reorientation, all the molecules of the same type (size and length of handles) are expected to align in the same direction while experiencing the same shear force.
This work describes a protocol to use such a shear flow to exert shear force on a massive set of molecular structures, as exemplified by the DNA i-motif. In this protocol, a shear flow is generated between the rotor and stator in a homogenizer tip. The present study found that the folded DNA i-motif structure could be unfolded by shear rates of 9724-97245 s−1. Besides, a dissociation constant of 36 µM was found between the L2H2-4OTD ligand and the i-motif. This value is consistent with that of 31 µM measured by the gel shift assay12. Further, the current technique is used to unfold the i-motif, which can expose the chelated copper (I) to catalyze a click reaction. This protocol thus allows one to unfold a large set of i-motif structures with low-cost instruments in a reasonable time (shorter than 30 min). Given that the shear force technique drastically increases the throughput of the force spectroscopy, we call this technique ensemble force spectroscopy (EFS). This protocol aims to provide experimental guidelines to facilitate the application of this shear force-based EFS.
Protocol
NOTE: All the buffers and the chemical reagents used in this protocol are listed in the Table Materials.
1. Preparation of the shear force microscope
NOTE: The shear force microscope contains two parts, a reaction unit (homogenizer) and a detection unit (fluorescence microscope). The magnification of the eyepiece is 10x, and the magnification of the objective lens (air) is 4x.
- Assemble the homogenizer and the microscope on a mounting table. Wear goggles and turn on the fluorescence microscope, and then adjust the homogenizer to make sure that the excitation light beam of an appropriate wavelength (here, 488 nm was used) passes through the center of the dispersing tip of the homogenizer.
- Prepare a flat-bottomed reaction chamber that is 5 cm in height and 1.5 cm2 x 1.5 cm2 in cross-section. To reduce the background, ensure that the selected chamber material does not fluoresce, such as glasses (some plastics have fluorescence).
- Choose an appropriate homogenizer dispersing tip that can provide the desired shear force.
NOTE: The shear rate is dependent on the distance between the rotor and stator at a fixed rotating speed11 according to the following equation:
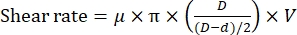
where µ is the dynamic viscosity of water at 20 °C; D is the inner diameter of the stator; d is the outer diameter of the rotor, and V is the shear speed (rpm/s). - Clamp the reaction chamber on the sample stage of the fluorescence microscope, and then adjust the chamber to hold the dispersing tip of the homogenizer (Figure 1). Make sure the dispersing tip is slightly above (~1 mm) the bottom surface of the reaction chamber.
- Adjust the vertical position of the homogenizer and the chamber together to ensure the focus of the microscope is on the surface of the dispersing tip. Then, adjust the horizontal position of the dispersing tip to ensure that the detection area (field of view) is set between the rotor and stator (Figure 1).
- Switch on the fluorescence channels according to the fluorescent dye used in the experiment.
- Before the high-speed shearing experiment, use deionized (DI) water to test the shearing with a low shearing speed (e.g., 2,000 rpm) to ensure the dispersing tip can work appropriately without touching the reaction chamber.
2. Unfolding i-motifs with and without ligands
- Prepare a human telomeric i-motif DNA (Table of Materials) labeled with a dye and a quencher at its two ends, respectively, in DI water, as described in Hu et al.11.
NOTE: i-motif containing sequence: 5'-TAA CCC TAA CCC TAA CCC TAA CCC TAA. - Dilute the DNA to 5 µM in the 30 mM MES buffer at pH 5.5 or pH 7.4. To the DNA solution, add ligand L2H2-4OTD, which was synthesized according to Abraham Punnoose et al.13, in a concentration range of 0-60 µM. Mix the solution gently for 10 min to fold the i-motif structures without light.
- Check and minimize the background fluorescence intensity of the reaction chamber that is filled with deionized DI water using the fluorescent microscope without shearing. An easy way to minimize the background fluorescence is to wash the reaction chamber with DI water. The background fluorescence value needs to be subtracted in the data analyses later.
NOTE: Stray light should be avoided from this step onwards. - Set the parameters of the CCD camera using the software. The recommended parameters are as follows: exposure time = 0.5 s, CCD sensitivity = 1600, and recording time = 20 min.
- Using a long pipet, add the DNA solution into the empty and clean reaction chamber. Cover the reaction chamber with a black box. Then, start the homogenizer to perform shearing at a selected shear rate ranging from 9,724 s−1 to 97,245 s−1 (selected using the software associated with the homogenizer) for 20 min with the CCD camera turned on to record the data.
- After the experiment, remove the chamber and wash it with DI water.
3. Shear force-actuated click reaction
- Prepare i-motif DNA in DI water. Incubate 10 µM i-motif DNA in 300 µL of 30 mM Tris buffer (pH 7.4) supplemented with 150 µM CuCl and 300 µM ascorbic acid for 10 min to fold the i-motif structures (all concentrations are final concentrations in the solution).
NOTE: CuCl is the catalyst of the click reaction. Ascorbic acid will prevent copper (I) oxidization. - Ultra-filtrate the solution with an ultrafiltration device at a centrifugal force of 14,300 x g. Replenish the solution to ~500 µL with 30 mM Tris buffer (pH 7.4) supplemented with 300 µM ascorbic acid after each filtration.
- Repeat the filtration 3x.
- Collect the residual solution and make a final volume of 300 µL by adding 30 mM Tris (pH 7.4) supplemented with 300 µM ascorbic acid along with 20 µM Calfluor 488 azide, 20 µM HPG, and 10 µM TBTA. Once the reagents are added, move the solution to the darkroom.
NOTE: Light should be avoided after this step. - Check and minimize the background fluorescence intensity of the reaction chamber filled with DI water using the microscope before the shearing experiments. An easy way to minimize the background fluorescence is to wash the reaction chamber with DI water.
- Add the DNA solution into the empty reaction chamber with a long pipet, and then start the homogenizer shearing at a shear rate of 63,209 s−1 for 20 min with the CCD camera turned on.
- After the experiment, remove the chamber and wash it with DI water.
Results
Figure 1 outlines the mechanical unfolding and real-time sensing of ensemble molecules in EFS. In Figure 1B, the fluorescence intensity of i-motif DNA was observed to increase with the shear rate ranging from 9,724 s−1 to 97,245 s−1 in a pH 5.5 MES buffer. As a control, fluorescence intensity was not increased when the same i-motif DNA was sheared at a rate of 63,209 s−1 in a pH 7.4 MES buffer. ...
Discussion
The protocol described in this manuscript allows real-time investigation of the unfolding of an ensemble set of biomolecular structures by shear force. The results presented here underscore that DNA i-motif structures can be unfolded by shear force. The unfolding of the ligand-bound i-motif and the shear force-actuated click reactions were proof-of-concept applications for this ensemble force spectroscopy method.
Figure 1 presents the instrument setup. The homogen...
Disclosures
The authors have no conflicts of interest.
Acknowledgements
This research work was supported by the National Science Foundation [CBET-1904921] and the National Institutes of Health [NIH R01CA236350] to H. M.
Materials
| Name | Company | Catalog Number | Comments |
| 3K MWCO Amicon | Millipore Sigma | ufc900324 | |
| Ascorbic acid | VWR | VWRC0143-100G | |
| Calfluor 488 azide | Click Chemistry Tools | 1369-1 | |
| CuCl | Thermo | ACRO270525000 | |
| Dispersion tip | Switzerland | PT-DA07/2EC-B101 | |
| DNA oligos | IDT | ||
| Dye | IDT | /5Cy5/ | |
| Fluorescence microscope | Janpan | Nikon TE2000-U | |
| Homogenizer | Switzerland | PT 3100D | |
| HPG | Santa Cruz Biotechnology | cs-295271 | |
| KCl | VWR | VWRC26760.295 | |
| MES | VWR | VWRCE169-500G | |
| Quencher | IDT | /3IAbRQSp/ | |
| TBTA | Tokyo Chemical Industry | T2993 | |
| Tris | VWR | VWRCE133-100G |
References
- Neuman, K. C., Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nature Methods. 5 (6), 491-505 (2008).
- Woodside, M. T., et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proceedings of the National Academy of Sciences of the United States of America. 103 (16), 6190-6195 (2006).
- Grandbois, M., Beyer, M., Rief, M., Clausen-Schaumann, H., Gaub, H. E. How strong is a covalent bond. Science. 283 (5408), 1727-1730 (1999).
- Strick, T. R., Allemand, J. F., Bensimon, D., Croquette, V. Behavior of supercoiled DNA. Biophysical Journal. 74 (4), 2016-2028 (1998).
- Yang, D., Ward, A., Halvorsen, K., Wong, W. P. Multiplexed single-molecule force spectroscopy using a centrifuge. Nature Communications. 7, 11026 (2016).
- Su, H., et al. Light-responsive polymer particles as force clamps for the mechanical unfolding of target molecules. Nano Letters. 18 (4), 2630-2636 (2018).
- Kirkness, M. W. H., Forde, N. R. Single-molecule assay for proteolytic susceptibility: Force-induced collagen destabilization. Biophysical Journal. 114 (3), 570-576 (2018).
- Astumian, R. D. Thermodynamics and kinetics of molecular motors. Biophysical Journal. 98 (11), 2401-2409 (2010).
- Bekard, I. B., Asimakis, P., Bertolini, J., Dunstan, D. E. The effects of shear flow on protein structure and function. Biopolymers. 95 (11), 733-745 (2011).
- Chistiakov, D. A., Orekhov, A. N., Bobryshev, Y. V. Effects of shear stress on endothelial cells: go with the flow. Acta Physiologica. 219 (2), 382-408 (2017).
- Hu, C., Jonchhe, S., Pokhrel, P., Karna, D., Mao, H. Mechanical unfolding of ensemble biomolecular structures by shear force. Chemical Science. 12 (30), 10159-10164 (2021).
- Sedghi Masoud, S., et al. Analysis of interactions between telomeric i-motif DNA and a cyclic tetraoxazole compound. ChemBioChem. 19 (21), 2268-2272 (2018).
- Abraham Punnoose, J., et al. Adaptive and specific recognition of telomeric G-quadruplexes via polyvalency induced unstacking of binding units. Journal of the American Chemical Society. 139 (22), 7476-7484 (2017).
- Dhakal, S., et al. Coexistence of an ILPR i-motif and a partially folded structure with comparable mechanical stability revealed at the single-molecule level. Journal of the American Chemical Society. 132 (26), 8991-8997 (2010).
- Hu, C., Tahir, R., Mao, H. Single-molecule mechanochemical sensing. Accounts of Chemical Research. 55 (9), 1214-1225 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved