A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Gene Digital Circuits Based on CRISPR-Cas Systems and Anti-CRISPR Proteins
* These authors contributed equally
In This Article
Summary
CRISPR-Cas systems and anti-CRISPR proteins were integrated into the scheme of Boolean gates in Saccharomyces cerevisiae. The new small logic circuits showed good performance and deepened the understanding of both dCas9/dCas12a-based transcription factors and the properties of anti-CRISPR proteins.
Abstract
Synthetic gene Boolean gates and digital circuits have a broad range of applications, from medical diagnostics to environmental care. The discovery of the CRISPR-Cas systems and their natural inhibitors-the anti-CRISPR proteins (Acrs)-provides a new tool to design and implement in vivo gene digital circuits. Here, we describe a protocol that follows the idea of the "Design-Build-Test-Learn" biological engineering cycle and makes use of dCas9/dCas12a together with their corresponding Acrs to establish small transcriptional networks, some of which behave like Boolean gates, in Saccharomyces cerevisiae. These results point out the properties of dCas9/dCas12a as transcription factors. In particular, to achieve maximal activation of gene expression, dSpCas9 needs to interact with an engineered scaffold RNA that collects multiple copies of the VP64 activation domain (AD). In contrast, dCas12a shall be fused, at the C terminus, with the strong VP64-p65-Rta (VPR) AD. Furthermore, the activity of both Cas proteins is not enhanced by increasing the amount of sgRNA/crRNA in the cell. This article also explains how to build Boolean gates based on the CRISPR-dCas-Acr interaction. The AcrIIA4 fused hormone-binding domain of the human estrogen receptor is the core of a NOT gate responsive to β-estradiol, whereas AcrVAs synthesized by the inducible GAL1 promoter permits to mimic both YES and NOT gates with galactose as an input. In the latter circuits, AcrVA5, together with dLbCas12a, showed the best logic behavior.
Introduction
In 2011, researchers proposed a computational method and developed a corresponding piece of software for the automatic design of digital synthetic gene circuits1. A user had to specify the number of inputs (three or four) and fill in the circuit truth table; this provided all the necessary information to derive the circuit structure using techniques from electronics. The truth table was translated into two Boolean formulae via the Karnaugh map method2. Each Boolean formula is made of clauses that describe logic operations (sum or multiplication) among (part of) the circuit inputs and their negations (the literals). Clauses, in their turn, are either summed up (OR) or multiplied (AND) to compute the circuit output. Every circuit can be realized according to any of its two corresponding formulae: one written in POS (product of sums) form and the other in SOP (sum of products) representation. The former consists of a multiplication of clauses (i.e., Boolean gates) that contain a logic sum of the literals. The latter, in contrast, is a sum of clauses where the literals are multiplied.
Electric circuits can be realized, on a breadboard, by physically wiring different gates together. The electric current permits the exchange of signals among gates, which leads to the computation of the output.
In biology, the situation is more complex. A Boolean gate can be realized as a transcription unit (TU; i.e., the sequence "promoter-coding region-terminator" inside eukaryotic cells), where transcription or translation (or both) are regulated. Thus, at least two kinds of molecules establish a biological wiring: the transcription factor proteins and the non-coding, antisense RNAs1.
A gene digital circuit is organized into two or three layers of gates, namely: 1) the input layer, which is made of YES (buffer) and NOT gates and converts the input chemicals into wiring molecules; 2) the internal layer, which consists of as many TUs as there are clauses in the corresponding Boolean formula. If the circuit is designed according to the SOP formula, every clause in the internal layer will produce the circuit output (e.g., fluorescence) in a so-called distributed output architecture. If the product of sum (POS) formula is used, then a 3) final layer is required, which will contain a single multiplicative gate collecting the wiring molecules from the internal layer.
Overall, in synthetic biology, many different schemes can be designed for the same circuit. They differ in the number and the kind of both TUs and wiring molecules. In order to choose the easiest solution to be implemented in yeast cells, each circuit design is associated with a complexity score S, defined as
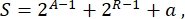
where A represents the number of activators, R represents the number of repressors, and a is the amount of antisense RNA molecules. If either activators or repressors are absent from the circuit, their contribution to S is zero. Therefore, it is more difficult to realize a circuit scheme in the lab (high S) when it requires a high number of orthogonal transcription factors. This means that new activators and repressors shall be engineered de novo in order to realize the complete wiring inside the digital circuits. In principle, novel DNA-binding proteins can be assembled by using Zinc Finger proteins3 and TAL effectors4 as templates. However, this option appears too arduous and time-consuming; therefore, one should rely mostly on small RNAs and translation regulation to finalize complex gene circuits.
Originally, this method was developed to fabricate digital circuits in bacteria. Indeed, in eukaryotic cells, instead of antisense RNAs, it is more suitable to talk of microRNAs (miRNAs) or small interfering RNAs (siRNAs)5. However, the RNAi pathway is not present in the yeast S. cerevisiae. Hence, one should opt for fully transcriptional networks. Suppose that a circuit needs five activators and five repressors; its complexity score would be S = 32. Circuit complexity can be reduced by replacing the 10 transcription factors with a single dCas96 (nuclease deficient Cas9) fused to an activation domain (AD). As shown in7, dCas9-AD works as a repressor in yeast when binding a promoter between the TATA box and the TSS (transcription start site) and as an activator when binding well upstream of the TATA box. Thus, one can replace 10 transcription factors with a single dCas9-AD fusion protein and 10 sgRNAs (single guide RNAs) for a total complexity score of S = 11. It is quick and easy to synthesize ten sgRNAs, whereas, as previously commented, the assembly of 10 proteins would demand much longer and more complicated work.
Alternatively, one might use two orthogonal dCas proteins (e.g., dCas9 and dCas12a): one to fuse to an AD, and the other bare or in combination with a repression domain. The complexity score would increase by only one unit (S = 12). Hence, CRISPR-dCas systems are the key to the construction of very intricate gene digital circuits in S. cerevisiae.
This paper deeply characterizes the efficiency of both dCas9- and dCas12a-based repressors and activators in yeast. Results show that they do not demand a high amount of sgRNA to optimize their activity, so episomal plasmids are preferentially avoided. Moreover, dCas9-based activators are far more effective when using a scaffold RNA (scRNA) that recruits copies of the VP64 AD. In contrast, dCas12a works well when fused to the strong VPR AD directly. Furthermore, a synthetic activated promoter demands a variable number of target sites, depending on the configuration of the activator (e.g., three when using dCas12a-VPR, six for dCas9-VP64, and only one with dCas9 and a scRNA). As a repressor, dCas12a appears more incisive when binding the coding region rather than the promoter.
As a drawback, however, CRISPR-dCas9/dCas12a do not interact with chemicals directly. Therefore, they might be of no use in the input layer. For this reason, alternative Boolean gate designs containing anti-CRISPR proteins (Acrs) have been investigated. Acrs act on (d)Cas proteins and inhibit their working8. Hence, they are a means to modulate the activity of CRISPR-(d)Cas systems. This paper thoroughly analyzes the interactions between type II Acrs and (d)Cas9, as well as type V Acrs and (d)Cas12a in S. cerevisiae. Since Acrs are much smaller than Cas proteins, a NOT gate responsive to the estrogen β-estradiol was built by fusing the hormone-binding domain of the human estrogen receptor9-HBD(hER)-to AcrIIA4. Besides, a handful of YES and NOT gates that expressed dCas12a(-AD) constitutively and AcrVAs upon induction with galactose were realized. At present, these gates serve only as a proof of concept. However, they also represent the first step toward a deep rethinking of the algorithm to carry out the computational automatic design of synthetic gene digital circuits in yeast cells.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Design and construction of the sgRNA/crRNA expression cassette
NOTE: There are two kinds of sgRNA/crRNA expression cassettes: one-termed SNR5210-is composed of the RNA polymerase III-dependent SNR52 promoter, the sgRNA/crRNA sequence, and the SUP4 terminator; another-abbreviated as RGR11-consists of the RNA polymerase II-dependent ADH1 promoter, the RGR (ribozyme-guide RNA-ribozyme) structure that contains two ribozymes (a hammerhead ribozyme-HH, and a hepatitis delta virus ribozyme-HDV) and the sequence of the sgRNA/crRNA in-between, and the ADH1 terminator. The sgRNA guiding Cas9 homologs are made of a spacer sequence and the characteristic direct repeat12, whereas the crRNA for Cas12a proteins comprises the direct repeat followed by the spacer sequence13,14 (see Supplementary Table 1 for all DNA sequences used in this study).
- Design the spacer sequence for Cas9/Cas12a-mediated transcriptional activation.
- Harness the bacterial lex operator sequence (named lexOp) to be the target site15,16 and insert it into the CYC1 promoter that drives the expression of the yeast-enhanced green fluorescent protein (yEGFP)17. Hence, the spacer sequence is defined by and complementary to the inserted lexOp.
- Check the orthogonality of the spacer sequence via the CRISPRDIRECT tool18.
- Paste the lexOp sequence flanked by the PAM sequence in the text field, define the PAM sequence as NRG for dCas9 and TTTV for dCas12a, and specify the species from the drop-down list as Budding Yeast (Saccharomyces cerevisiae) S288C Genome. Click on Design. Make sure there is no matching target site in 20mer+PAM nor in 12mer+PAM search.
- Construct the sgRNA/crRNA expression cassette.
- Use touchdown PCR to amplify the DNA sequences of standard biological parts, like promoters, coding sequences, and terminators.
- Prepare a reaction mixture containing: 20-40 ng of DNA template, 1 µL of 10 µM forward primer (i.e., ot25, sgRNA/crRNA expression plasmid construction), 1 µL of 10 µM reverse primer (i.e., ot26, sgRNA/crRNA expression plasmid construction), 5 µL of 2.5 mM dNTP mix, 0.5 µL of DNA polymerase, 10 µL of 5x DNA polymerase reaction buffer, and double distilled water (ddH2O) up to a total volume of 50 µL.
NOTE: Refer to Supplementary Table 2 for a list of primers used in this study. - Run the touchdown PCR program on a thermocycler:
Stage 1: 98 °C for 30 s.
Stage 2 with 10 cycles: 98 °C for 10 s, 68 °C for 20 s, and 72 °C for 15 s.
Stage 3 with 25 cycles: 98 °C for 10 s, 59 °C for 20 s, and 72 °C for 15 s.
Stage 4: 72 °C for 2 min.
Finally, hold at 4 °C until starting the follow-up experiments.
NOTE: The 68 °C in stage 2 and 59 °C in stage 3 depend on the melting temperatures of both forward and reverse primers, varying from different pairs of primers. The time at 72 °C in stages 2 and 3 is determined by the PCR product's length and the DNA polymerase's speed.
- Prepare a reaction mixture containing: 20-40 ng of DNA template, 1 µL of 10 µM forward primer (i.e., ot25, sgRNA/crRNA expression plasmid construction), 1 µL of 10 µM reverse primer (i.e., ot26, sgRNA/crRNA expression plasmid construction), 5 µL of 2.5 mM dNTP mix, 0.5 µL of DNA polymerase, 10 µL of 5x DNA polymerase reaction buffer, and double distilled water (ddH2O) up to a total volume of 50 µL.
- Isolate the PCR products via gel electrophoresis (100 V, 30 min). Elute the DNA sequences from the agarose gel via a DNA gel extraction kit (see Table of Materials).
NOTE: A 0.8% agarose gel is required for fragments longer than 500 nt, a 1.5% agarose gel for fragments between 100 nt and 500 nt, and a 2% agarose gel for fragments shorter than 100 nt. - Insert the TU expressing sgRNA/crRNA into a pRSII404/424 shuttle vector19, which contains the ampicillin resistance gene and the yeast-selectable auxotrophic marker gene-TRP1.
- Digest the shuttle vector at 37 °C for 1 h with the two restriction enzymes SacI and Acc65I. Prepare the reaction mixture by adding 5 µg of the shuttle vector, the recommended amounts of enzymes, digestion buffer (according to the enzyme instructions), and ddH2O up to a total volume of 30 µL.
- Verify the shuttle vector digestion via gel electrophoresis (see substep 1.3.2). Then, inactivate the two enzymes at 65 °C for 20 min.
- Use the Gibson assembly method20 to insert the purified PCR products into the cut-open shuttle vector by letting in the equimolar DNA mixture at 50 °C for 1 h.
- Transform Escherichia coli DH5α competent cells with the above Gibson reaction mixture via the heat shock transformation method21. Transfer the transformed E. coli cells onto Luria-Bertani (LB) agar plates containing ampicillin (0.1 g/L). Put the plates into the incubator at 37 °C and let the cells grow overnight.
- Pick four colonies from the LB agar plate and culture them separately overnight at 37 °C in LB solution containing ampicillin (0.1 g/L). Then, use the mini-preparation procedure to extract plasmids from the E. coli cells22.
- Use the primers ot18 and ot19 (see Supplementary Table 2 for oligo sequences) to sequence and confirm the inserted transcription unit via the Sanger method23.
NOTE: In later experiments, the constructed and confirmed plasmids will be inserted into the yeast genome via the PEG/LiAc protocol24.
- Use touchdown PCR to amplify the DNA sequences of standard biological parts, like promoters, coding sequences, and terminators.
2. Design and construction of the scaffold RNA expression cassette
NOTE: The scaffold guide RNA (scRNA) is composed of the sgRNA sequence and the MS2 hairpin structures25. Two kinds of MS2 hairpin structures are used in this work: wild-type MS2 hairpin-wt, and f6 MS2 coat protein (MCP) aptamer-f6.
- Synthesize an scRNA template that is able to accommodate different spacer sequences (i.e., pSNR52-spacer_DR(SpCas9)-2×MS2(wt+f6)-SUP4t).
NOTE: In this study, the scRNA template was synthesized by a gene synthesis company (see Table of Materials). - Design proper primers (see Supplementary Table 2) to run PCR on the necessary spacer sequences.
- Follow the procedures in step 1.3 to construct an scRNA expression cassette.
3. dSpCas9 engineering and expression plasmid construction
- Obtain the plasmid pTPGI_dSpCas9_VP64 (see Table of Materials).
- Construct the acceptor vector pRSII406-pGPD-ATG-XbaI-XhoI-CYC1t, based on the pRSII406 shuttle vector, via touchdown PCR and the Gibson assembly method (refer to step 1.3). The plasmid provides a strong constitutive promoter-pGPD, and a terminator-CYC1t.
- Digest the pTPGI_dCas9_VP64 plasmid and the newly constructed acceptor vector (5 µg for 1 h or 10 µg for overnight-see step 1.3.3.1 as a reference) with XbaI and XhoI at 37 °C. Separate and purify the insert fragment and acceptor vector as in step 1.3.2.
- Ligate the purified insert fragment and the acceptor vector with T4 DNA ligase at 16 °C for 8 h. Prepare the ligation solution by adding 50-100 ng of the purified acceptor vector, purified target fragments in equimolar amount with the acceptor vector, 1.5 µL of T4 buffer, 0.5 µL of T4 ligase, and ddH2O up to a total volume of 15 µL.
- Follow steps 1.3.3.3 and 1.3.3.4. Then, confirm that the newly constructed plasmid is correct by digestion with XbaI and Xhol (37 °C, 1 h) and gel electrophoresis (see step 1.3.2).
4. dCas12a engineering and plasmid construction
- Construct the plasmids expressing dCas12a-AD.
- Synthesize two yeast codon-optimized dCas12a proteins (denAsCas12a and dLbCas12a) flanked by BamHI and Xhol restriction enzyme sites.
NOTE: In this study, the two yeast codon-optimized dCas12a proteins were synthesized by a gene synthesis company (see Table of Materials). - Construct the acceptor vector pRSII406-promoter-ATG-NLS-GS-HIStag-GS-BamHI-sp-XhoI-AD-NLS-TAA-mTGUO1 via touchdown PCR and the Gibson assembly method (see step 1.3), where the "promoter" is either pGPD or pGAL1, "sp" represents a short random sequence, and "AD" is either VPR or VP64.
- Insert each dCas12a protein into the two newly constructed acceptor vectors via digestion with BamHI and XhoI and ligation with T4 DNA ligase (refer to steps 3.3 and 3.4).
- Synthesize two yeast codon-optimized dCas12a proteins (denAsCas12a and dLbCas12a) flanked by BamHI and Xhol restriction enzyme sites.
- Construct the plasmids expressing a bare dCas12a.
- Construct an acceptor vector for the galactose-inducible expression cassettes of dCas12a as pRSII406-Acc651-pGAL1-ATG-NLS-GS-HIStag-GS-BamHI-sp-XhoI-GS-NLS-TAA-CYC1t using touchdown PCR and the Gibson assembly method (see step 1.3).
- Digest the plasmids containing the dCas12a proteins and the above acceptor vector with BamHI and Xhol, then ligate them with T4 DNA ligase to get the plasmid pRSII406-pGAL1-Acc651-ATG-NLS-GS-HIStag-GS-BamHI-dCas12a-XhoI-GS-NLS-TAA-CYC1t (refer to steps 3.3 and 3.4).
- Digest the plasmid obtained in step 4.2.2 with Acc65I and BamHI, and then replace pGAL1 with pGPD via touchdown PCR and the Gibson assembly method to construct pRSII406-pGPD-ATG-NLS-GS-HIStag-GS-BamHI-dCas12a-XhoI-GS-NLS-TAA-CYC1t.
5. Anti-CRISPR protein engineering and plasmid construction
NOTE: Three kinds of promoters have been employed to drive Acrs expression: an inducible promoter-pGAL1, four yeast constitutive promoters-pGPD, pACT1, pTEF1, and pTEF2, and a synthetic constitutive promoter-genCYC1t_pCYC1noTATA26.
- Obtain the plasmids containing the sequences of type II Acrs (AcrIIA2, AcrIIA427, and AcrIIA528) and type V-A Acrs (AcrVA1, AcrVA4, and AcrVA529) from a gene synthesis company.
- Construct the plasmids based on the pRSII403 shuttle vector to express Acrs.
- Construct AcrIIA expression cassettes.
NOTE: Use touchdown PCR to amplify four different promoters (i.e., pGPD, pACT1, pTEF2, and genCYC1t_pCYC1noTATA), the three kinds of AcrIIA, and two terminators (ADH1t and CYC1t). Build a series of TUs expressing AcrIIAs, under different promoters, via the Gibson assembly method (refer to step 1.3). - Construct AcrVA expression cassettes.
- Synthesize the acceptor sequence: ATG-FLAGtag-GS-BamHI-sp-XhoI-NLS-GS-NLS-TAA-Tsynth6, where "sp" is a random sequence that will be replaced by AcrVAs later.
NOTE: In this study, the acceptor sequences were synthesized by a gene synthesis company (Table of Materials). - Assemble the acceptor vectors pRSII403-promoter-ATG-FLAGtag-GS-BamHI-sp-XhoI-NLS-GS-NLS-TAA-Tsynth6, where the "promoter" is: pGAL1, pTEF1, and genCYC1t_pCYC1noTATA. Use the Gibson assembly method (refer to step 1.3).
- Insert each of the three AcrVAs into the acceptor vector described in step 5.2.2.2 (via touchdown PCR and the Gibson assembly method [see step 1.3]) to build a set of plasmids producing AcrVAs.
- Synthesize the acceptor sequence: ATG-FLAGtag-GS-BamHI-sp-XhoI-NLS-GS-NLS-TAA-Tsynth6, where "sp" is a random sequence that will be replaced by AcrVAs later.
- Construct AcrIIA expression cassettes.
- Further engineer AcrIIA4 by extending its sequence with the sequences of the GS linker and HBD(hER). This permits the building of a β-estradiol-responsive circuit.
- Use touchdown PCR to obtain GS-HBD and AcrIIA4 parts separately (see step 1.3.1).
- Put AcrIIA4, GS-HBD, and the shuttle vector into the Gibson mixture and construct the complete plasmid via the Gibson method (see step 1.3.3).
6. crRNA detection: RT-qPCR and primers' design
NOTE: crRNA detection was achieved via RT-qPCR, which is organized in three steps.
- Perform the RNA extraction and purification from yeast cells via an RNA kit.
- Culture yeast cells overnight in 2 mL of synthetic defined complete medium (SDC, 1 L volume: 20 g of Glucose, 2 g of AA mix, 6.7 g of YNB, 396 mg of Leucine, 79.2 mg of Tryptophan, 79.2 mg of Histidine, and 79.2 mg of Uracil) by using a 24-well plate (240 rpm, 30 °C).
- In the morning, dilute the cell culture (1:100) into 2 mL of fresh SDC and keep growing the yeast cells at 30 °C, 240 rpm, for another 4 h.
- Harvest the whole 2 mL of the cell solution and centrifuge at 20,238 x g for 2 min. Remove the supernatant carefully since the cell pellet is small.
- Extract the RNA from yeast cells using an RNA kit.
- Check the RNA quality.
- Prepare a 1% agarose gel. Mix 5 µL of each RNA sample with 1 µL of DNA loading dye. Then load the mixture on the gel and run it.
- Ensure that two clear bands at ~4,000 nt and ~2,000 nt, corresponding to ribosomal RNA (25S/18S), are present on the gel. A further blurred band can be seen at ~80 nt for tRNA.
- Use the RNA samples immediately for cDNA synthesis or store them at -80 °C for future use.
- Reverse transcription: Use the stem-loop method30 to form the first strand of cDNA corresponding to the crRNA (40 nt). For the reverse transcription of the sgRNA (almost 100 nt), the procedure is the same as that for the cDNA synthesis of the reference gene ACT1.
NOTE: The method for the reverse transcription of the crRNA is different from that used with the sgRNA and the ACT1 mRNA. Since the crRNA is very short, it was treated as a microRNA and used the microRNA reverse transcription method (stem-loop approach) to obtain the corresponding cDNA. Two cDNA synthesis kits (a stem-loop kit for the crRNA and a usual reverse transcription kit for the ACT1 gene) were used in crRNA quantification. The same amount of RNA was used in the two kits (see Table of Materials) to make the experiment results comparable. The primer used with the stem-loop kit was designed according to the stem-loop sequence and the last six nucleotides at the 3' end of the crRNA (for the stem-loop and primer sequence, see Supplementary Table 2).- Stem-loop method for crRNA reverse transcription
- Take the RNA template, primer, and buffers out of the freezer and let them melt on ice.
- Genomic DNA removal: First, prepare 10 µL of the reaction mixture according to the kit instructions. Put the mixture in a thermal cycler at 42 °C for 2 min. Finally, transfer it onto ice.
- Synthesis of the first cDNA strand: Prepare 20 µL of the reaction mixture by adding 10 µL of the mixture from step 6.2.1.2, 1 µL of stem-loop primer (2 µM concentration), 2 µL of 10x RT reaction buffer, 2 µL of reverse transcription enzyme mix (containing the reverse transcriptase), and 5 µL of RNase-free H2O.
- Place the reaction mixture in a thermal cycler and run the following program: 25 °C for 5 min, 50 °C for 15 min, and 85 °C for 5 min. Use the product for the qPCR reaction immediately or store it at -80 °C.
- sgRNA and ACT1 mRNA reverse transcription
- First reaction: Prepare a 13 µL mixture consisting of the primer mix, dNTP mix, RNA template (50 pg-5 µg), and RNase-free water (apart from the RNA template, all provided by the kit), according to the kit instructions. Use 1 µg of RNA template. Put the mixture into a thermal cycler at 70 °C for 10 min.
- Second reaction (cDNA synthesis): Prepare the reaction mixture by adding the reagents (as described in the kit instructions) to the 13 µL of the first reaction solution up to a final volume of 20 µL. Place the mixture in a thermal cycler for 15 min at 50 °C and then for an additional 5 min at 85 °C. Use the product for the qPCR reaction immediately or store it at -80 °C.
- Stem-loop method for crRNA reverse transcription
- SYBR kit for qPCR: Ct value detection
NOTE: The reverse primer used in the qPCR of the crRNA is fixed because it corresponds to the reverse complement of the stem-loop sequence (see Supplementary Table 2). The forward primer, in contrast, is variable and depends on the sequence of the crRNA. The forward and reverse primers for the sgRNA and ACT1 mRNA qPCR are designed at https://www.ncbi.nlm.nih.gov/tools/primer-blast/. Two primers are selected when the difference between their melting temperatures is no greater than 2 °C (see Supplementary Table 2). Each sample is measured in three replicates.- Prepare the qPCR reaction mixture according to the manufacturer's instructions for the SYBR kit.
- Set the following qPCR program in a real-time PCR machine.
Preincubation: 10 min at 95 °C.
PCR stage: 15 s at 95 °C, followed by 34 s at 55 °C. Set the cycle of the PCR stage to 45 times. Melting stage: 10 s at 95 °C, followed by 60 s at 65 °C, and finally 1 s at 97 °C. - Calculate the relative mRNA expression values via the Pfaffl formula31.
7. Immunofluorescence to detect Cas proteins
NOTE: Cas proteins (CasP) are fused to a His_tag.
- Yeast cell preparation
- Pick some yeast cells using a sterile loop and culture them in 5 mL of YPD-rich medium overnight at 240 rpm at 30 °C. In the morning, add 500 µL of the cell solution to 20 mL of fresh YPD and grow them at 240 rpm at 30 °C until OD600 reaches 0.6.
- Take 5 mL of the culture and mix it with 500 µL of 37% formaldehyde. Let the mixture stay at room temperature (RT) for 10 min. Harvest the cells by centrifugation at 1,500 x g for 5 min.
- Remove the supernatant and resuspend the cells with 1 mL of fixation buffer (0.1 M KH2PO4, 0.5 M MgCl2, 3.7% formaldehyde, pH = 6.5). Keep the cell solution at RT for 20 min.
- Centrifuge the cell solution at 1,500 x g for 5 min. Discard the supernatant and resuspend the cells in 1 mL of washing buffer (0.1 M KH2PO4, 1.2 M sorbitol, pH = 6.5) supplemented with 4 µL of beta-mercaptoethanol and 4 µL of lysate solution (5 mg/mL). Put the cell solution into an incubator at 37 °C for 20 min.
- Centrifuge the cell solution at 1,500 x g for 5 min and discard the supernatant. Wash the cell pellet twice with 1 mL of PBS by centrifugation (1,500 x g for 5 min).
- Resuspend the cells in 100 µL of PBS plus 0.05% Tween 20 and add 4 µL of BSA solution (10 mg/mL). Keep the cell solution at RT for 20 min.
- Incubation with primary antibody
- Add the Anti-His tag primary antibody to the mixture in step 7.1.6 at a 1:400 dilution. Keep the solution at RT for 2 h.
- Centrifuge the mixture in step 7.2.1 at 1,500 x g for 5 min and remove the supernatant. Add 1 mL of PBST and centrifuge (1,500 x g) for 5 min to wash the cell pellet. Repeat this operation twice. Finally, discard the supernatant and suspend the cells in 100 µL of 1x PBST.
- Microscopy cell detection
- Mount the cells on a slide; take 2 µL of the cell solution from step 7.2.2 and place it on a glass slide. Cover it with a coverslip.
- Observe the cells under a fluorescence microscope. Turn on the fluorescent light source, microscope, and computer. Write down the fluorescent light source number and open the microscope software on the computer.
- Put the slide on the microscope stage. Choose the 40x objective lens and observe the cells under the green light (550 nm). Move the coarse focus knob until the contour of yeast cells appears. Move the fine focus knob to focus the cells.
- Detect the cells with the microscope software. Close the microscope field of view and switch to the computer screen. Click on Live, wait for 3-5 s, and click on Capture to take a picture. Save the picture.
- Turn off the computer, microscope, and fluorescent light source.
8. Data acquisition: FACS
NOTE: Green fluorescence is detected via flow cytometry (i.e., fluorescence-activated cell sorting [FACS] measurements). Yeast cells are cultured, in general, at 30 °C and 240 rpm to run FACS experiments. However, cells might demand some precautions depending on their genetic content. Cells that contain the dCas12a-VPR gene (controlled by the GPD constitutive promoter) must be grown for 24 h in the SDC solution. Afterward, cells are diluted at a ratio of 1:100 in fresh SDC and grown for another 12 h before measuring the fluorescence intensity. Cells modified with the AcrIIA4-HBD(hER) gene demand dilution as well. Moreover, OD600 needs to be controlled. First, the cells are allowed to grow in SDC overnight (over 14 h). In the morning, OD600 is measured. Then the culture is diluted in SDC, supplied with a diverse concentration of β-estradiol, down to OD600 = 0.1. Before FACS experiments, the cells are grown for another 7 h such that OD600 reaches 0.8-1.0. Cells expressing dCas9-VP64 or dCas12a-VP64 are grown in SDC for 20-24 h without dilution and further growth before the measurements at the FACS machine.
- Switch on the FACS machine 20 min before the measurements to warm up the laser.
- Prepare the samples (dilution): mix 20 µL of the cell culture with 300 µL of ddH2O.
- Run the FACS software on the computer connected to the FACS machine and create a new experiment. Set the measurement parameters (i.e., FSC(SSC)-A/H/W and histogram).
- Select the filter according to the excitation and emission wavelengths of the samples. For example, the target protein here is yEGFP, whose excitation and emission wavelengths are 488 nm and 507 nm, respectively. So, select the FITC or GFP filter (excitation wavelength: 488 nm; emission wavelength: 527/32 nm). Set the acquisition cell number to 10,000.
- Adjust the FITC filter voltage by measuring the intensity of fluorescent beads. Ensure that the relative difference in the intensity of the beads between two consecutive experiments does not exceed 5%.
- Wash the machine with ddH2O for a few seconds to remove any possible excess beads.
- Measure the sample fluorescence intensity. Click on Preview and wait for 3-5 s for sample injection stability. Finally, click on Acquire.
- Measure the beads again at the end of the experiment. The voltage is the one that was used during the initial beads' measurement (see step 8.4, 438-441 V). Check whether the relative difference between the two beads' measurements exceeds 5%.
- Export the FACS data as FCS files.
- Analyze the FCS files with R Studio software.
9. Data analysis
NOTE: Use the Flowcore R Bioconductor package32 within R studio. The FCS files were analyzed using a script written in R language.
- Open R studio and load the script Bdverse analysis.R to analyze the FCS files.
- Set the experiment name (ename), the directory (path) where the FCS files are
Stored (dir_d), and where the result files will be created (dir_r). - Set the fluorescence channel. For instance, select_ch = "GPA-A" if green fluorescence was measured.
- Set the number of samples that were measured (s_num).
- Set the dimensions of each dot plot (sxlim, sylim). Set the maximal length of the x and y axis for bar plots and box plots (xlimit, ylimit). xlimit must be greater than or equal to s_num.
- Choose the gating method by removing the # from the corresponding lines.
NOTE: morphGate is an automatic gating method carried out by the script (i.e., the region of the dot plots where the cells are denser is recognized and selected by the program). polygonGate and rectangleGate demand to look at the dot plots and define either the vertices of a polygon or the side of a rectangle that embraces the zone where most of the cells lie. - Select the flowSet object gated, corresponding to the chosen gating method. Select the dot plot resolution (res; should be at least equal to 256).
- Uncomment flt_low <- filter_low(sp) to remove measurements where fluorescence is negative. Uncomment flt_sp <- filter(flt_lw) to remove outliers due to other experiments.
- Press Source and run the script. All the files containing the results from the analysis are created in dir_r.
Access restricted. Please log in or start a trial to view this content.
Results
sgRNA/crRNA expression by an RNA polymerase III-type promoter
First, this work addressed the engineering of the transcriptional activation circuit (circuit 1) shown in Figure 1A. It contained three basic components: 1) the gene encoding for yEGFP (the reporter), which was preceded by a series of different synthetic promoters that provided target sites for dCas9/dCas12a-AD; 2) a yeast codon-optimized version of dCas9 or dCas12a fused to an activation domain (VP64 and VP...
Access restricted. Please log in or start a trial to view this content.
Discussion
The protocol showed a possible complete workflow for synthetic gene digital circuits, following the "Design-Build-Test-Learn" (DBTL) biological engineering cycle and concerning both dry-lab and wet-lab experiments. Here, we focused on the CRISPR-Cas system, mainly dSpCas9, denAsCas12a, dLbCas12a, and the corresponding anti-CRISPR proteins, by designing and building in S. cerevisiae small transcriptional networks. Some of them mimicked Boolean gates, which are the basic components of digital circuits. All...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors declare no competing financial interest.
Acknowledgements
We want to thank all the students of the Synthetic Biology lab-SPST, TJU-for their general help, together with Zhi Li and Xiangyang Zhang for their assistance in FACS experiments.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 0.1 mL PCR 8-strip tubes | NEST | 403112 | |
| 0.2 mL PCR tubes | Axygen | PCR-02-C | |
| 1.5 mL Microtubes | Axygen | MCT-150-C | |
| 15 mL Centrifuge tubes | BIOFIL | CFT011150 | |
| 2 mL Microtubes | Axygen | MCT-200-C | |
| 50 mL Centrifuge tubes | BIOFIL | CFT011500 | |
| Agarose-molecular biology grade | Invitrogen | 75510-019 | |
| Ampicillin sodium salt | Solarbio | 69-52-3 | |
| Applied biosystems veriti 96-well thermal cycler | Thermo Fisher Scientific | 4375786 | |
| AxyPrep DNA gel extraction kit | Axygen | AP-GX-250 | |
| BD FACSuite CS&T research beads | BD | 650621 | Fluorescent beads |
| BD FACSVerse flow cytometer | BD | - | |
| Centrifuge | Eppendorf | 5424 | |
| Centrifuge Sorvall ST 16R | Thermo Fisher Scientific | 75004380 | |
| E. coli competent cells (Strain DH5α) | Life Technologies | 18263-012 | |
| ECL select Western Blotting detection reagent | GE Healthcare | RPN2235 | |
| Electrophoresis apparatus | Beijing JUNYI Electrophoresis Co., Ltd | JY300C | |
| Flat 8-strip caps | NEST | 406012 | |
| Gene synthesis company | Azenta Life Sciences | https://web.azenta.com/zh-cn/azenta-life-sciences | |
| Goat anti-Mouse IgG (H+L) cross-adsorbed secondary antibody Alexa Fluor 568 | Invitrogen | A-11004 | |
| HiFiScript cDNA synthesis kit | CWBIO | CW2569M | Kit used in step 6.2.2.1 |
| Lysate solution (Zymolyase) | zymoresearch | E1004-A | |
| Nikon Eclipse 80i fluorescence microscope | Nikon | - | Fluorescence microscope |
| Pipet tips—10 μL | Axygen | T-300-R-S | |
| Pipet tips—1000 μL | Axygen | T-1000-B-R-S | |
| Pipet tips—200 μL | Axygen | T-200-Y-R-S | |
| pRSII403 | Addgene | 35436 | |
| pRSII404 | Addgene | 35438 | |
| pRSII405 | Addgene | 35440 | |
| pRSII406 | Addgene | 35442 | |
| pRSII424 | Addgene | 35466 | |
| pTPGI_dSpCas9_VP64 | Addgene | 49013 | |
| Q5 High-fidelity DNApolymerase | New England Biolabs | M0491 | |
| Restriction enzyme-Acc65I | New England Biolabs | R0599 | |
| Restriction enzyme-BamHI | New England Biolabs | R0136 | |
| Restriction enzyme-SacI-HF | New England Biolabs | R3156 | |
| Restriction enzyme-XhoI | New England Biolabs | R0146 | |
| Roche LightCycler 96 | Roche | - | Real-Time PCR Instrument |
| S. cerevisiae CEN.PK2-1C | - | - | The parent strain. The genotype is: MATa; his3D1; leu2-3_112; ura3-52; trp1-289; MAL2-8c; SUC2 |
| Stem-Loop Kit | SparkJade | AG0502 | Kit used in step 6.2.1.3 |
| T100 Thermal Cycler | BIO-RAD | 186-1096 | |
| T4 DNA ligase | New England Biolabs | M0202 | |
| T5 Exonuclease | New England Biolabs | M0363 | |
| Taq DNA ligase | New England Biolabs | M0208 | |
| Taq DNA polymerase | New England Biolabs | M0495 | |
| TB Green Premix Ex Taq II (Tli RNaseH Plus)(2x) (SYBR Green I dye) | Takara | RR820Q | |
| YeaStar RNA kit | Zymo Research | R1002 | |
| β-estradiol | Sigma-Aldrich | E8875 |
References
- Marchisio, M. A., Stelling, J. Automatic design of digital synthetic gene circuits. PLOS Computational Biology. 7 (2), 1001083(2011).
- Karnaugh, M. The map method for synthesis of combinational logic circuits. Transactions of the American Institute of Electrical Engineers. 72 (9), 593-599 (1953).
- Mandell, J. G., Barbas, C. F. Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Research. 34, 516-523 (2006).
- Bogdanove, A. J., Voytas, D. F. TAL effectors: customizable proteins for DNA targeting. Science. 333 (6051), 1843-1846 (2011).
- Drinnenberg, I. A., et al. RNAi in budding yeast. Science. 326 (5952), 544-550 (2009).
- Gander, M. W., Vrana, J. D., Voje, W. E., Carothers, J. M., Klavins, E. Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nature Communications. 8, 15459(2017).
- Farzadfard, F., Perli, S. D., Lu, T. K. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synthetic Biology. 2 (10), 604-613 (2013).
- Nakamura, M., et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nature Communications. 10 (1), 194(2019).
- Louvion, J. F., Havaux-Copf, B., Picard, D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 131 (1), 129-134 (1993).
- DiCarlo, J. E., et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Research. 41 (7), 4336-4343 (2013).
- Gao, Y., Zhao, Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. Journal of Integrative Plant Biology. 56 (4), 343-349 (2014).
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816-821 (2012).
- Zetsche, B., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163 (3), 759-771 (2015).
- Fonfara, I., Richter, H., Bratovic, M., Le Rhun, A., Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 532 (7600), 517-521 (2016).
- Yu, L., Marchisio, M. A. Saccharomyces cerevisiae synthetic transcriptional networks harnessing dCas12a and Type V-A anti-CRISPR proteins. ACS Synthetic Biology. 10 (4), 870-883 (2021).
- Zhang, Y., Marchisio, M. A. Interaction of bare dSpCas9, scaffold gRNA, and type II anti-CRISPR proteins highly favors the control of gene expression in the yeast S. cerevisiae. ACS Synthetic Biology. 11 (1), 176-190 (2022).
- Sheff, M. A., Thorn, K. S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21 (8), 661-670 (2004).
- Naito, Y., Hino, K., Bono, H., Ui-Tei, K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 31 (7), 1120-1123 (2015).
- Chee, M. K., Haase, S. B. New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomyces cerevisiae. G3: Genes|Genomes|Genetics. 2 (5), 515-526 (2012).
- Gibson, D. G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Research. 37 (20), 6984-6990 (2009).
- Froger, A., Hall, J. E. Transformation of plasmid DNA into E. coli using the heat shock method. Journal of Visualized Experiments. (6), e253(2007).
- Green, M. R., Sambrook, J. Molecular Cloning. Fourth edition. , Cold Spring Harbor Laboratory Press. NY. (2012).
- Sanger, F. Determination of nucleotide sequences in DNA. Science. 214 (4526), 1205-1210 (1981).
- Gietz, R. D., Woods, R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in Enzymology. 350, 87-96 (2002).
- Zalatan, J. G., et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 160 (1-2), 339-350 (2015).
- Song, W., Li, J., Liang, Q., Marchisio, M. A. Can terminators be used as insulators into yeast synthetic gene circuits. Journal of Biological Engineering. 10, 19(2016).
- Rauch, B. J., et al. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell. 168 (1-2), 150-158 (2017).
- Hynes, A. P., et al. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nature Microbiology. 2 (10), 1374-1380 (2017).
- Watters, K. E., Fellmann, C., Bai, H. B., Ren, S. M., Doudna, J. A. Systematic discovery of natural CRISPR-Cas12a inhibitors. Science. 362 (6411), 236-239 (2018).
- Chen, C., et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research. 33 (20), 179(2005).
- Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 29 (9), 45(2001).
- Hahne, F., et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 10 (1), 106(2009).
- Li, J., Xu, Z., Chupalov, A., Marchisio, M. A. Anti-CRISPR-based biosensors in the yeast S. cerevisiae. Journal of Biological Engineering. 12, 11(2018).
- Dong, L., et al. An anti-CRISPR protein disables type V Cas12a by acetylation. Nature Structural & Molecular Biology. 26 (4), 308-314 (2019).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved