Methods Article
Imaging ATG9A, a Multi-Spanning Membrane Protein
In This Article
Summary
This protocol describes various methods that can help in the study of ATG9A biology, including immunofluorescence followed by image analysis, transient overexpression considerations, and investigating the ATG9A glycosylation status using western blot.
Abstract
Autophagy is a highly conserved pathway that the cell uses to maintain homeostasis, degrade damaged organelles, combat invading pathogens, and survive pathological conditions. A set of proteins, called ATG proteins, comprise the core autophagy machinery and work together in a defined hierarchy. Studies in recent years have improved our knowledge of the autophagy pathway. Most recently, it has been proposed that ATG9A vesicles are at the heart of autophagy, as they control the rapid de novo synthesis of an organelle called the phagophore. The study of ATG9A has proven challenging, since ATG9A is a transmembrane protein, and it is present in different membrane compartments. As such, understanding its trafficking is an important element for understanding autophagy. Here, detailed methods are presented that can be used to study ATG9A and, in particular, its localization using immunofluorescence techniques, which can be assessed and quantified. The pitfalls of transient overexpression are also addressed. The correct characterization of ATG9A function and the standardization of techniques to analyze its trafficking are crucial to further characterize the events governing autophagy initiation.
Introduction
ATG9A is the only transmembrane protein of the core autophagy machinery and is trafficked between the Golgi and a cytosolic ATG9A vesicle compartment, transiting through the endosomal compartment1. Having long been enigmatic, ATG9A has recently been described to function as a lipid scramblase, as it equilibrates lipids across membrane bilayers2,3. It is now clear that ATG9A resides at the top of the hierarchy in autophagosome formation, and its study is, thus, vital for understanding autophagy4,5. As such, ATG9A vesicles have been recently proposed as the "seed" of the autophagosome6,7. However, previous studies have demonstrated that ATG9A only transiently interacts with the forming autophagosome at different steps of its maturation and does not integrate into the autophagic membrane6,8,9,10,11. Thus, further investigations are needed to completely unravel the role and potential multiple functions of ATG9A in autophagosome formation. However, the discrepancy between the current models and the previous data can only be resolved through targeted experiments addressing the trafficking of ATG9A using validated quantitative approaches and intracellular markers.
There are various tools in use to study ATG9A, each with advantages and disadvantages, and the use of these tools is complicated by the structure of ATG9A, its molecular function, and cellular trafficking2,8,12. ATG9A forms a homotrimer, is glycosylated, and is trafficked throughout the cell to compartments such as the Golgi, the endosomes, and the plasma membrane13,14. Given its complex itinerary, there are several challenges in interpreting readouts such as ATG9A dispersal from the Golgi upon specific treatments or stimuli (such as nutrient and serum starvation). ATG9A is extremely dynamic in terms of vesicular trafficking; indeed, ATG9A-containing vesicles have been defined as the ATG9A compartment in the context of starvation-induced autophagy. The ATG9A compartment, formed by these dynamic vesicles, transiently interacts with several intracellular organelles8,15,16,17. The techniques described here, including immunofluorescence, live imaging, and glycosylation assays, should aid in the detection and understanding of ATG9A biology. In particular, the approaches described in this article will help to address questions about localization to specific cellular compartments and interactions with specific protein partners and/or membrane compartments. As the ATG9A hydrophobic conserved core domain (PFAM domain PF04109) has a unique topology and ATG9A cycles between several membrane compartments, researchers should be aware of certain pitfalls and artifacts when transiently overexpressing ATG9A, including, but not restricted to, endoplasmic reticulum (ER) retention. Other possible issues may arise due to misfolding of the protein, artifactual aggregation in normal growing conditions, or insufficient detection of the vesicular compartment due to suboptimal permeabilization protocols for immunofluorescence.
When imaging endogenous ATG9A, care must be taken in the sample preparation and image acquisition to ensure the quality of the subsequent quantitative analysis and the correct interpretation of the data. Combining the techniques described in this article with standard biochemical approaches (such as immunoprecipitation or pull-down experiments not described here) should improve our understanding of ATG9A function. This experimental toolkit is intended to help new researchers navigate some of the assays required to determine the function of ATG9A in their biological system.
Protocol
All the reagents used in this study are commercially available, except for the ATG9A DNA constructs and homemade STO-215 antibody (see Table of Materials), which are available upon request. The analysis tools described here are based on open-source software (FIJI/ImageJ)18.
1. Cell culture
- Maintain HEK293A cells in a T150 tissue culture treated flask to 80%-90% confluence in high-glucose DMEM (Dulbecco's Modified Eagle Medium) supplemented with 10% FBS (fetal bovine serum) and 4 mM L-glutamine (see Table of Materials). Incubate the cells at 37 °C in a humidified tissue culture incubator at 10% CO2.
NOTE: HEK293A cells are used in the present study, as they have a robust canonical autophagy response upon amino acid starvation, which is, in particular, detected by an increase in lipidated, membrane-associated LC31,9,19, and are suitable for imaging. - Passage the cells by aspirating the medium from the flask using a serological pipette. Wash the cells once with 15 mL of 1x PBS (phosphate-buffered saline) or a similar solution before adding 2 mL of Trypsin-EDTA (ethylenediaminetetraacetic acid) solution to detach the cells. Collect the detached cells with 8 mL of DMEM, and seed back a number of cells so that 80%-90% confluency is reached after 2 days for the experiments described here.
2. Endogenous staining of ATG9A
- Seed HEK293A cells (or cells of choice) on sterile No. 1.5 glass coverslips placed into a 24-well plate so that they are ~80% confluent the next day. This typically gives 7 x 104 cells/well in 500 μL of DMEM.
NOTE: For HEK293A cells or other loosely adherent cell lines, the coverslips need to be coated with poly-D-Lysine at 0.1 mg/mL in deionized water. Add 500 μL of the poly-D-lysine (see Table of Materials) on the top of the coverslips placed into the wells for 10 min at room temperature (RT), followed by three washes with deionized water and a last wash in DMEM. Proceed to seeding the cells after the coating, and take care that they are evenly spread out in the culture dish. - Treat the cells according to the specific experimental conditions (i.e., starvation for 2 h in EBSS [Earle's Balanced Salt Solution]).
NOTE: The EBSS composition is 1 g/L D-glucose, 6.8 g/L NaCl, 0.4 g/L KCl, 0.151 g/L CaCl2.2H2O, 0.2 g/L mM MgSO4.7H2O, 0.124 g/L NaH2HPO4.2H2O, and 2.2 g/L NaHCO3 (see Table of Materials) dissolved in distilled water. - Aspirate the medium, and carefully replace it with 500 μL of 4% formaldehyde solution in PBS supplemented with 0.1 mM CaCl2 and 0.1 mM MgCl2 for 20 min at RT to fix the cells.
- Aspirate the 4% formaldehyde solution from each well, replacing it with 500 μL of PBS. Repeat this wash step three times. Do not let the coverslips dry out or remain without liquid.
- Quench the free aldehyde groups by using 500 μL of 50 mM NH4Cl solution in PBS for 10 min at RT.
- Aspirate the PBS, and replace it with 500 μL of a 50 μg/mL digitonin solution in PBS (stock solution 1 mg/mL in deionized water, see Table of Materials) for 5 min at RT to permeabilize the cells.
- Aspirate the digitonin solution from each well, and replace it with 500 μL of PBS. Repeat this wash step three times.
- Aspirate the PBS from each well, and replace it with 500 μL of blocking solution (5% BSA [bovine serum albumin] diluted in PBS) for 30 min at RT.
- Aspirate the blocking solution, and replace it with 500 μL of PBS.
- Using tweezers, collect the coverslips from the well, carefully remove the excess PBS solution using thin tissue wipes or cellulose filter paper, and gently lay down each coverslip, cell side down, onto a 50 μL drop of primary antibody solution (e.g., Armenian Hamster 14F2 diluted to 0.9 μg/mL in 1% BSA/PBS solution, see Table of Materials). Incubate in a humidified chamber for 1 h at RT.
NOTE: For easy handling, the drops of antibody solution can be placed on a sheet of self-sealing thermoplastic film (see Table of Materials) within another container instead of directly onto a solid surface. - Collect the coverslips using tweezers, and, after gently draining off the excess primary antibody solution using thin tissue wipes, replace the coverslips (cell side up) in the 24-well plate, and wash them three times with PBS.
- Repeat step 2.10. Using tweezers, collect the coverslips from each well, carefully draining off the excess PBS using thin tissue wipes, and gently lay down each coverslip (cell side down) onto a 50 μL drop of secondary antibody solution diluted 1:1,000 in 1% BSA/PBS solution (e.g., Cy3 Goat Anti-Armenian Hamster IgG, which has minimal crossreactivity with bovine, human, mouse, rabbit, and rat serum proteins, see Table of Materials). Incubate in a humidified chamber for 1 h at RT.
NOTE: Optional: A cytoskeleton marker can be used in addition to the secondary antibody for the subsequent image analysis (i.e., Alexa Fluor 647 Phalloidin diluted 1:1,000, see Table of Materials). For easy handling, the drops of antibody solution can be placed on a sheet of sealing film within another container instead of directly onto a solid surface. - Collect the coverslips using tweezers and, after draining off the excess secondary antibody solution, place the coverslips in the 24-well plate (cell side up) and wash them three times with 500 μL PBS.
- Using tweezers, collect the coverslips, carefully draining off the excess PBS using thin tissue wipes, and gently lay down each coverslip (cell side down) onto a 50 μL drop of 1:4,000 Hoechst solution (Hoechst 33342) in PBS. Incubate in a humidity chamber for 5 min at RT.
NOTE: For easy handling, the drops of antibody solution can be placed on a sheet of sealing film within another container instead of directly onto a solid surface. - Collect the coverslips using tweezers, and, after gently draining off the excess Hoechst solution using thin tissue wipes, replace the coverslips in the 24-well plate, and wash three times with PBS and once with de-ionized water (500 μL per wash).
- Carefully drain the excess deionized water using thin tissue wipes, and gently lay down each coverslip (cell side down) onto a 10-20 μL drop of mounting solution (see Table of Materials) spotted onto a microscope slide for immunofluorescence, avoiding the formation of air bubbles.
NOTE: The mounting medium used here has the same refractive index as immersion oil and hardens after a few hours at RT or overnight at 4 °C. Non-hardening mounting solutions can be used, but care must be taken to seal the coverslip with nail polish. - Remove the excess mounting solution by aspiration, and let the samples dry while lying flat at RT overnight in the dark, either in a slide holder or covered with aluminum foil.
3. Image acquisition
- Turn on the confocal microscope. Open the imaging software (see Table of Materials) to start the image acquisition setup.
- In the Function tab, select the Plan-Apochromat 63x/1.4 Oil DIC M27 objective lens to capture the images for analysis.
- In the Acquisition Function tab, turn on the appropriate lasers in the Laser zone of the Control Panel (Argon, Diode-405-30, DPSS 561-10, and HeNe633).
- In the Imaging Setup zone of the Control Panel, create four tracks, with each track corresponding to one channel, to perform sequential acquisition.
- Set the appropriate resolution of the images in the Acquisition Mode window. Set the resolution at 1,024 x 1,024 (Frame size), and select a Bit depth of 16-bit.
- For each channel, adjust the laser power and gain to obtain a good signal without saturated pixels, which would impede the image intensity analysis. Using the Live button, set the Laser output level and Gain (Master).
NOTE: The use of the range indicator option is recommended for saturated pixel detection. Keep the laser power between 1% and 10% and the Gain (Master) below 850 to avoid background noise. - In the Channels zone of the Control Panel, set the same pinhole aperture for each channel, considering 1 Airy Unit (AU) for the channel with the highest wavelength.
- Acquire 10 random fields containing similar amounts of cells (20-30 cells per field). The acquisition of 100-200 cells per condition will provide good power for the later image analysis.
4. Image analysis of ATG9A dispersal
- Download FIJI software from the internet (see Table of Materials). Open the image with FIJI by clicking on Plugins > Bio-Formats > BioFormats Importer.
- Click on Analyse > Set measurements, and select the Mean gray value in the measurements window for the intensity analysis.
- Click on Image > Color > Split Channels, and separate the channels into three images (Golgi marker, cytoskeleton, ATG9A signal).
- Go to Image > Adjust > Threshold, and define a threshold in the channel corresponding to the Golgi marker.
- Click on Edit > Selection > Create Selection, and create a selection from the binary image.
- Go to Edit > Selection > Add to Manager > Rename (Golgi), save the selection in the ROI manager, and rename it to Golgi.
- Click on Image > Adjust > Threshold, and define a threshold in the channel corresponding to the cytoskeleton marker to define the cell contour.
- Go to Edit > Selection > Create Selection, and create a selection from the binary image.
- Click on Edit > Selection > Add to Manager > Rename (Total), save the selection in the ROI manager, and rename it Total.
- Select the image corresponding to the ATG9A staining.
- In Analyse > Measure, apply the ROI Golgi by clicking on it, and measure the intensity of the Golgi region.
- Click on Analyse > Measure, apply the ROI Total by clicking on it, and measure the intensity of the Total region.
- Repeat the procedure in all the images, save the results as .csv files, and process the data using the following formula:
ATG9A dispersal rate = Golgi intensity/Total intensity
5. Live cell imaging of ATG9A constructs
- Seed HEK293A cells (or cells of choice) in 2 mL of medium into a 60 mm tissue culture dish so they reach ~65%-70% confluency the next day; this typically gives 1 x 106-2 x 106 cells.
- The following day, prepare Lipofectamine:DNA mixes (or a suitable alternative DNA transfection reagent, see Table of Materials) as follows:
- Depending on construct expression efficiency, dilute 0.5-2 µg of plasmid DNA into 100 µL of a suitable serum-free medium, and gently mix the solution by pipetting up and down. Incubate the mixture for 5 min at RT.
NOTE: The plasmid DNA construct is experiment-specific. Researchers may either clone by themselves or obtain from the authors upon request. - Dilute Lipofectamine 2000 transfection reagent at a ratio of 3:1 Lipofectamine:DNA into 100 µL of a suitable serum-free medium and gently mix the solution by pipetting up and down. Incubate this mix for 5 min at RT.
- Mix both solutions together by gently pipetting up and down and incubating for 20 min at RT.
- Depending on construct expression efficiency, dilute 0.5-2 µg of plasmid DNA into 100 µL of a suitable serum-free medium, and gently mix the solution by pipetting up and down. Incubate the mixture for 5 min at RT.
- Add the Lipofectamine:DNA mix to each cell culture plate containing 4 mL of growth medium, and gently rock the plate back and forth to distribute the mix evenly. Incubate the cells at 37 °C in a humidified cell culture incubator at 10% CO2.
- Replace the medium with fresh growth medium 4 h after transfection, and incubate the cells at 37 °C in a humidified cell culture incubator at 10% CO2 overnight.
- The next day, trypsinize, count, and reseed the cells on culture dishes suitable for live-cell microscopy (culture dishes with a No. 1.5 glass coverslip, see Table of Materials). Seed 0.4 x 106-0.7 x 106 cells onto the dishes to reach ~60%-75% confluency on the coverslip at the time of imaging.
NOTE: Coating with poly-D-lysine, as explained in step 2.1, is recommended for HEK293A cells. - The next day (48 h after transfection), image the cells.
6. Investigating the glycosylation state of ATG9A
- Seed HEK293A cells (or cells of choice) in a 10 cm tissue culture dish so they reach ~80% confluency the next day if planning to investigate endogenous ATG9A, which typically gives 1.5 x 106-2.5 x 106 cells in 10 mL. Alternatively, seed the cells so they reach ~65%-70% confluency if transfecting ATG9A constructs, which typically gives 1 x 106-1.5 x 106 cells in 10 mL.
- Treat the cells with 100 µg/mL cyclohexamide (CHX, see Table of Materials) or vehicle the next day. Incubate for 24 h (or until a desired time point).
- Remove the cells from the incubator, aspirate the medium, and place on ice. Replace it with 5 mL of ice-cold 1x PBS. Physically detach the cells from the dish using a cell scraper, and pipette the PBS-cell solution into a 15 mL conical-bottom tube. Centrifuge at 800 x g for 5 min at 4 °C to pellet the cells.
- Aspirate the supernatant, and resuspend the cells in ~100 µL (depending on the size of the cell pellet) of ice-cold TNTE lysis buffer (1% Triton, 150 mM NaCl, 20 mM Tris-HCl, pH 7.4, 0.5 mM EDTA) supplemented with protease inhibitor cocktail tablets (see Table of Materials), and transfer to a 1.5 mL microcentrifuge tube.
- Incubate the lysate on ice for 15 min. After this, centrifuge the lysate at 20,000 x g for 10 min at 4 °C to sediment the nuclei and insoluble debris, and transfer the supernatant to a fresh microcentrifuge tube.
- Quantify the protein concentration of the lysate using the Bradford method20 and a spectrophotometer that can measure at a wavelength of 595 nm.
- Normalize the protein amounts, and combine 16 µL of the lysate (containing no more than 100 µg of protein in total) with 5x PNGase F (Peptide:N-glycosidase F) buffer according to the manufacturer's instructions before adding 1 µL of PNGase F enzyme (see Table of Materials). Incubate as instructed by the PNGase F manufacturer.
- Add a volume of 3x Laemmli sample buffer to achieve a 1x concentration, and incubate at 65 °C for 5 min before loading for electrophoresis using a Tris-acetate gel to maximize the separation of proteins. Transfer the proteins from the gel to a suitable membrane (i.e., PVDF [polyvinylidene difluoride]) using standard western blot protocols21 (see Table of Materials).
NOTE: Boiling the samples at 95 °C will cause ATG9A to be aggregated, thus reducing the detection of ATG9A. - Perform the western blot using specific antibodies for ATG9A (STO-215 antibody, produced in-house1) (see Table of Materials). Leave the higher-molecular weight section of the membrane uncut to visualize the higher ATG9A molecular weight species.
Results
ATG9A is a transmembrane protein associated with several intracellular membrane compartments8,17,22,23,24. In basal conditions, ATG9A is mainly localized at the trans-Golgi network (TGN), as indicated by the immunofluorescence of the endogenous protein and the overlaps with GM130, a cis-Golgi marker (Figure 1A), as well as in small vesicles that partially overlap with the endocytic recycling compartment (ERC)23. ATG9A localization at the Golgi can be detected using different immunofluorescence protocols. However, the vesicular fraction of ATG9A, as well as its change of localization, in particular the increase in the vesicular pool, in response to specific stimuli such as nutrient and serum starvation, can be quite variable in intensity and difficult to visualize with conventional imaging approaches. The ratio between ATG9A localized at the Golgi and ATG9A localized to a vesicular fraction is termed the ATG9A dispersal rate. To detect changes in the ATG9A dispersal rate, for instance upon EBSS treatment, which is used to deplete both serum and amino acids, a Golgi marker such as GM130 or TGN46 and a cytoskeleton marker such as Phalloidin, which stains the cell contour25, are useful to readily quantify the ATG9A dispersal (Figure 1B). Importantly, the mean fluorescence ratio analysis can only be interpreted as a comparative measure between conditions rather than as a fixed rate of dispersal. The ratio between compartments is highly dependent on biological and non-biological factors such as the cell line used, the staining quality, or the thresholding methods applied (Figure 1B). For this reason, the researcher needs to set up a pipeline that is able to detect ATG9A Golgi enrichment in their specific experimental conditions and then extend the analysis with the same parameters to all the images in the set to be analyzed. Representative binary images and areas selected for the analysis of ATG9A mean fluorescence are shown as a guide in Figure 1B.
ATG9A harbors several transmembrane domains flanked by two relatively flexible and unstructured N- and C-terminal domains, of which the C-terminal sequence encompasses almost half the protein12. Importantly, the localization pattern of overexpressed ATG9A can be influenced by which protein end is tagged (Figure 2A). In particular, when using transient expression systems and tagging ATG9A directly on its N-terminus with a fluorescent tag (e.g., eGFP, mRFP, or derivatives), its Golgi localization can be partially compromised, with less enrichment seen in basal (i.e., fed) conditions, while the ATG9A vesicles are still readily visible (Figure 2A). Tagging ATG9A on its C-terminus seems to slightly induce larger GFP positive clusters that could be aggregated. Finally, a monomeric version of mRFP-ATG9A also shows similar fluorescent clusters of vesicles and little Golgi staining in overexpressing cells (Figure 2A).
ATG9A folds in the ER membrane before being trafficked to the Golgi and ATG9A vesicles. During its residence in the ER, ATG9A becomes modified by N-linked glycans on Asparagine 99, and then upon reaching the Golgi, it acquires complex, mature N-linked glycans1,14. This modification by glycosylation can be detected through western blot by the appearance of a double band14. Consistent with its intracellular localization, most endogenous ATG9A harbors complex N-linked glycans, and, therefore, the higher-molecular weight band is predominant, with a faint lower-molecular weight band also visible (Figure 2B). The presence of a double band is most readily seen when using Tris-acetate gels to improve the resolution of higher-molecular weight proteins (Figure 2B, control, t = 0). When the endogenous protein is subjected to PNGase F (Peptide:N-glycosidase F) treatment, which removes most of the complex N-linked glycans, the protein runs as a single band (Figure 2B, PNGase F, t = 0). Therefore, the N-linked glycosylation status of ATG9A can be used as a proxy to monitor the exiting of ATG9A from the ER to the Golgi, which is reflected by the relative ratio between the two bands.
When transfecting mRFP-ATG9A constructs transiently, the overexpressed protein initially accumulates in the ER, potentially because the trafficking machinery is unable to fold and traffic all the ATG9A, and the lower molecular weight band is predominant (Figure 2C, control t = 0). Notably, after 24 h of expression of mRFP-ATG9A, there is approximately an equal distribution between the upper and lower bands, suggesting that the mRFP-ATG9A pool is moving into the Golgi (Figure 2C, Control, t = 24). If the cells are treated with cycloheximide (CHX), which blocks de novo protein synthesis26, the folding and exit of ATG9A from the ER can be clarified. As the endogenous protein is folded, glycosylated, and resident in the Golgi, treatment with CHX does not significantly alter the ratio of the lower- and higher-molecular weight bands (Figure 2B, Control). However, using the transient expression of mRFP-ATG9A, the CHX treatment promotes the accumulation of the higher-molecular weight band (Figure 2C, Control, CHX t = 24). The higher-molecular weight overexpressed mRFP-ATG9A band collapses into the lower band after treatment with PNGase F (Figure 2C, PNGase F, t = 24). These data show that the endogenous protein rapidly acquires mature glycans, as reflected by the predominance of the higher-molecular weight band, and the CHX chase does not affect the ratio of the double bands (Figure 2B). In the case of transiently overexpressed mRFP-ATG9A, CHX treatment induces the accumulation of the upper band, indicating that more mature glycans are acquired as the ER pool folds and exits the ER to the Golgi (Figure 2C).
The addition of a linker between the ATG9A sequence and the fluorescent tags can be helpful in promoting a more physiological localization and trafficking of the protein. Fusing a 3x-FLAG sequence (24 amino acids) between an N-terminal fluorophore and ATG9A helps the overexpressed protein behave similarly to the endogenous one (Figure 3). Indeed, overexpressed mCherry-3xFLAG-ATG9A colocalizes with the Golgi marker GM130 in fed conditions (Figure 3A). Importantly, this localization and the ATG9A vesicular compartment are preserved over time, allowing the spatiotemporal study of the trafficking of ATG9A (Figure 3B).
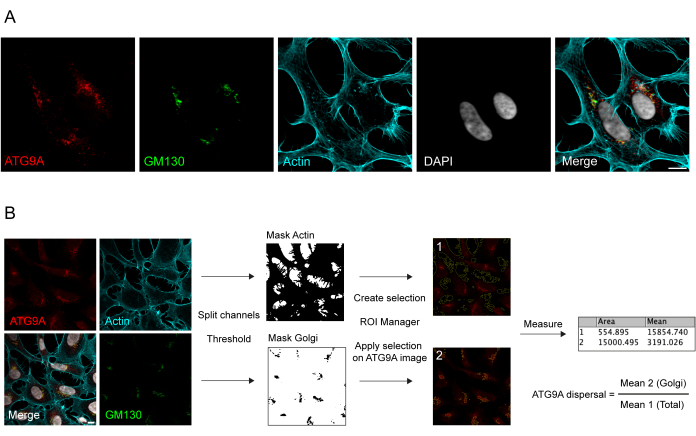
Figure 1: Image analysis of endogenous ATG9A localization. (A) Representative immunofluorescence image of endogenous ATG9A (red), GM130 as a Golgi marker (green), and Phalloidin to visualize the actin cytoskeleton (cyan). Scale bar = 10 μm. (B) Workflow of the image analysis to determine the fraction of endogenous ATG9A that localizes at the Golgi area. Scale bar = 10 μm. Please click here to view a larger version of this figure.
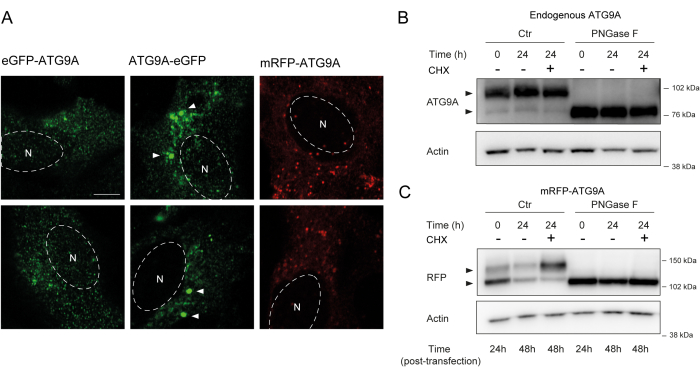
Figure 2: Analysis of fluorescently tagged-ATG9A constructs by localization and glycosylation. (A) eGFP N-terminally tagged ATG9A is less localized at the Golgi and primarily resides in the vesicles. eGFP C-terminally tagged ATG9A exhibits aggregates within the cell (some examples are marked by white arrowheads; the eGFP-ATG9A and ATG9A-eGFP are in green). mRFP N-terminally tagged ATG9A is less localized at the Golgi and primarily resides in the vesicles. N denotes the approximate location of the cell nucleus, and the mRFP-ATG9A is in red. Scale bar = 5 μm. (B) Endogenous ATG9A appears as two bands when analyzed by western blot (arrowheads): an upper band (complex N-linked glycans) and a lower band (no mature N-linked glycans). Treatment with cyclohexamide (CHX) does not affect the ratio between the upper and lower bands. Treatment with PNGase F causes the disappearance of the upper band. (C) After transient transfection of mRFP-tagged ATG9A in HEK293A cells, two prominent bands are visible on western blot (arrowheads). Treatment with PNGase F causes the disappearance of the upper band. Treatment with CHX after transfection leads to increased glycosylation as the pool of transfected ATG9A is trafficked from the ER to the Golgi. Please click here to view a larger version of this figure.
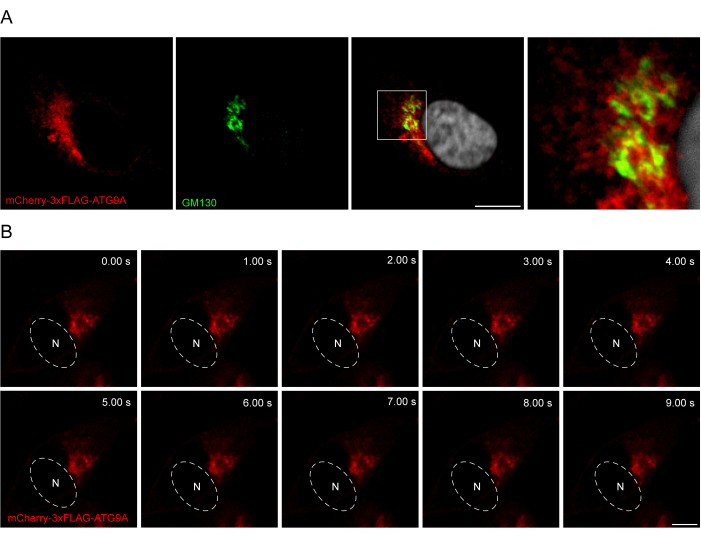
Figure 3: Analysis of mCherry-3xFLAG-ATG9A localization by immunofluorescence and live imaging. (A) Immunofluorescence experiments of HEK293A cells transiently overexpressing mCherry-3xFLAG-ATG9A and stained with the Golgi marker GM130. Scale bar = 10 μm. The mCherry-3xFLAG-ATG9A is in red, and the GM130 Golgi marker is in green. (B) Montage from live-imaging experiments in HEK293A cells transiently overexpressing mCherry-3xFLAG-ATG9A. N denotes the approximate location of the nucleus. Time frame = 1 fps. Scale bar = 10 μm. The mCherry-3xFLAG-ATG9A is in red. Please click here to view a larger version of this figure.
Discussion
This study illustrates the various tools that can be used to investigate ATG9A localization. Firstly, this study describes how ATG9A can be visualized by immunofluorescence and how this can be quantified. Secondly, strategies that can be used to tag ATG9A with a fluorescent marker for visualization in either fixed or live cells are compared. Finally, this work describes how to investigate and use the glycosylation state of ATG9A to determine if ATG9A has exited the ER and trafficked through the Golgi.
Regarding the characterization of endogenous ATG9A localization by immunofluorescence, care must be taken with the fixation and permeabilization methods employed for the experiment. According to the standard procedures here described, paraformaldehyde fixation coupled with digitonin permeabilization are good conditions to visualize both Golgi-associated ATG9A and ATG9A-positive vesicles7. Together with fixation and permeabilization, the timing of incubation with the primary antibody solution is also critical. We have observed, but not documented, that higher concentrations of primary antibody solution and longer incubation times can lead to a misrepresentative increase in the Golgi staining of ATG9A, which eventually compromises the detection of ATG9A redistribution to other membrane compartments. Additionally, since ATG9A is present in many intracellular compartments1,13,17,22,23,24,27,28,it is important to use specific membrane markers, together with ATG9A, to identify where ATG9A is located. Several approaches have been used in the past to quantify ATG9A localization, including Pearson's correlation coefficient for colocalization29. However, the partial overlap of ATG9A with the Golgi and the distinct vesicular compartment leads to a high number of pixel outliers, which may bias the interpretation of the correlation coefficient. For this reason, a more simplistic approach based on the ratio of the mean fluorescence in the two compartments to be analyzed is preferred, and this approach is less sensitive to cell-by-cell variability. For further information on image analysis through microscopy, readers are directed to this book chapter30.
When investigating the glycosylation status of ATG9A, the selection of gels for running the western blots is important. For this protocol, 3%-8% Tris-acetate gels are preferred because they offer the highest resolution for larger proteins, but alternative gel compositions or running buffers that offer a good separation of high-molecular weight proteins can also be used. The experimenter can ensure the maximal separation of proteins by increasing the time of electrophoresis.
When preparing the samples to visualize ATG9A on western blot, care should be taken not to boil the samples after adding the Laemmli buffer; boiling at 95 °C induces the formation of ATG9A aggregates, and subsequently, ATG9A does not migrate efficiently into the gel1. Heating the samples at 65 °C for 5 min is recommended27.
High levels of transfection usually lead to higher accumulation of ATG9A in the ER, while moderate expression levels help the physiological localization of the protein. Anecdotally, incubation times of 72 h instead of 48 h often help reduce ER localization artifacts. Notably, mRFP-ATG9A can accurately report on ATG9A trafficking and function if the levels are controlled through expression levels or by using stable cell lines8,9,22,27.
The failure of a population of overexpressed ATG9A to acquire mature N-linked glycans can be used as a read-out for perturbed ATG9A trafficking. When mutating or deleting certain regions of ATG9A, there is a risk of increased ER retention, which can lead to failure to acquire mature N-linked glycans and, thus, a faster-migrating ATG9A band on western blot. Researchers working with truncated ATG9A constructs should check for ER retention, glycosylation states, and Golgi localization.
For the live-cell imaging of ATG9A, an Airyscan microscope, relying on the fast Airyscan function, provides optimal resolution of typically about 120 nm. For localization accuracy, frame rates of around 1-2 frames per second (fps) in super-resolution mode are optimal depending on how many channels are imaged. Similar confocal microscopes that can image at high speed can also be used for the imaging of ATG9A vesicles; however, it should be noted that the imaging speed can directly affect the detection of events and, therefore, affect the interpretation of the data.
In summary, the presented protocols describe ways to quantify and characterize ATG9A localization by immunofluorescence, live-cell microscopy, and its glycosylation status. These protocols can aid researchers working with ATG9A and help avoid some pitfalls.
Disclosures
S.A.T. serves on the scientific advisory board of Casma Therapeutics.
Acknowledgements
The authors thank Rocco D'Antuono for proofreading aspects of the manuscript, as well as all present and past members of the Molecular Cell Biology of Autophagy (MCBA) lab for the discussions that led to the refinement of these protocols. A. v.V., S.d.T., E.A., S.A.T, were supported by The Francis Crick Institute which receives its core funding from Cancer Research UK (CC2134), the UK Medical Research Council (CC2134). This research was funded in whole, or in part, by the Wellcome Trust (CC2134). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Materials
| Name | Company | Catalog Number | Comments |
| 24 multiwell plates | Falcon | 353047 | For tissue culture |
| 35 mm Dish | No. 1.5 Coverslip | 14 mm Glass Diameter | Uncoated | MATTEK | P35G-1.5-14-C | Cell culture dish for live-cell microscopy |
| 4x Laemmli Sample Buffer | Bio-Rad | 1610747 | |
| 60 mm tissue culture dish | Thermofischer Scientific | 10099170 | For tissue culture |
| Alexa Fluor 647 Phalloidin | Thermofischer Scientific | A22287 | Actin stain |
| anti-ATG9A antibody | home made | STO-215 | Rabbit anti N-terminal peptide ATG9A |
| anti-Rabbit IgG, peroxidase-linked | Invitrogen | 10794347/NA934-1ml | Secondary antibody for rabbit polyclonal STO-215 |
| anti-RFP antibody | Evrogen | AB233 | for Western Blot |
| ATG9A Monoclonal Antibody (14F2 8B1), Invitrogen | Invitrogen | 15232826 | Antibody for immunofluorescence |
| ATG9A-eGFP | home made | Construct which expresses tagged ATG9A | |
| Bemis Parafilm | Thermofischer Scientific | 11747487 | self-sealing thermoplastic film |
| Bio-Rad Protein Assay Dye Reagent Concentrate | Bio-Rad | 5000006 | For determining protein concentration |
| Bovine serum albumin (BSA) | Merck | 10735086001 | For blocking non-specific labelling |
| CaCl2.2H2O | / | For PBS and EBSS | |
| cOmplete Protease Inhibitor Cocktail | Roche | 11697498001 | Supplement in lysis buffer to prevent protein degradation |
| Cy3 AffiniPure Goat Anti-Armenian Hamster IgG | Jackson ImmunoResearch | 127-165-099 | Secondary antibody for i14F2 8B1 antibody for mmunofluorescence |
| Cyclohexamide | Sigma Aldrich | 66-81-9 | To stop protein translation |
| D-Glucose | / | For EBSS | |
| Digitonin | Merck | 300410 | For permeabilizing cells |
| DMEM | Merck | D6546-6x500ml | For tissue culture |
| eGFP-ATG9A | home made | Construct which expresses tagged ATG9A | |
| Fetal Bovine Serum | Gibco | 10270-106 | Supplement for DMEM for cell culture |
| FIJI (ImageJ) | / | https:/fiji.sc/ | Open source image analysis software |
| Hoechst | Thermofischer Scientific | H3570 | Stains the nucleus |
| KCl | / | For EBSS | |
| L-glutamine | Sigma | 67513 | For tissue culture |
| Lipofectamine 2000 | Invitrogen | 11668-019 | For Cell Transfection |
| LSM880 Airyscan microscope | Zeiss | / | Confocal microscopy |
| MgCl2 | / | For PBS | |
| MgSO4.7H2O | / | For EBSS | |
| Mowiol mounting solution | Millipore | 475904 | for permanent mounting glass coverslips |
| NaCl | / | For EBSS | |
| NaH2PO4.2H2O | / | For EBSS | |
| NaHCO3 | / | For EBSS | |
| NuPAGE 3 to 8%, Tris-Acetate, 1.5 mm, Mini Protein Gels | Thermofischer Scientific | EA0378BOX | for Western Blotting |
| NuPAGE MES SDS Running Buffer (20x) | Life Tech | NP0002 | for Western Blotting |
| Opti-MEM I Reduced Serum Medium | Thermo | 31985062 | For Cell Transfection |
| Paraformaldehyde | Agar Scientific | R1026 | For fixing cells |
| pcDNA3.1-mCherry-3xFlag-ATG9A | home made | Construct which expresses tagged ATG9A | |
| Phosphate Buffered Saline (PBS) | / | For tissue culture | |
| PNGaseF | NEB | P0710S | To remove N-linked glycans |
| Poly-D-lysine hydrobromide mol wt 70,000-150,000 | Merck | P0899 | For coating coverslips |
| Rapid PNGase F enzyme | NEB | P07105 | To remove N-linked glycans |
| RFP-ATG9A | home made | Construct which expresses tagged ATG9A | |
| Triton X-100 | Thermofischer Scientific | 13454259 | Detergent for Cell lysis |
| Trypsin-EDTA solution | Sigma | T4049 | For tissue culture |
| Whatman Filter Paper | Merck | WHA1001325 | For Western Blot and IF |
| XCell SureLock Mini-Cell Electrophoresis System | Invitrogen | EI0001 | For Western Blotting |
| Zen Black edition | Zeiss | / | Used to operate the LSM 880 |
References
- Young, A. R., et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. Journal of Cell Science. 119, 3888-3900 (2006).
- Maeda, S., et al. lipid scrambling activity and role in autophagosome formation of ATG9A. Nature Structural & Molecular Biology. 27 (12), 1194-1201 (2020).
- Matoba, K., et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nature Structural & Molecular Biology. 27 (12), 1185-1193 (2020).
- Mercer, T. J., Gubas, A., Tooze, S. A. A molecular perspective of mammalian autophagosome biogenesis. Journal of Biological Chemistry. 293 (15), 5386-5395 (2018).
- Yamamoto, H., Zhang, S., Mizushima, N. Autophagy genes in biology and disease. Nature Reviews: Genetics. , (2023).
- Sawa-Makarska, J., et al. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science. 369 (6508), (2020).
- Melia, T. J., Lystad, A. H., Simonsen, A. Autophagosome biogenesis: From membrane growth to closure. Journal of Cell Biology. 219 (6), 202002085 (2020).
- Orsi, A., et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Molecular Biology of the Cell. 23 (10), 1860-1873 (2012).
- Judith, D., et al. ATG9A shapes the forming autophagosome through Arfaptin 2 and phosphatidylinositol 4-kinase IIIbeta. Journal of Cell Biology. 218 (5), 1634-1652 (2019).
- Karanasios, E., et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nature Communications. 7, 12420 (2016).
- Koyama-Honda, I., Itakura, E., Fujiwara, T. K., Mizushima, N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 9 (10), 1491-1499 (2013).
- Guardia, C. M., et al. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Reports. 31 (13), 107837 (2020).
- Soreng, K., et al. SNX18 regulates ATG9A trafficking from recycling endosomes by recruiting Dynamin-2. EMBO Reports. 19 (4), e44837 (2018).
- Staudt, C., Gilis, F., Boonen, M., Jadot, M. Molecular determinants that mediate the sorting of human ATG9A from the endoplasmic reticulum. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1863 (9), 2299-2310 (2016).
- Knaevelsrud, H., Carlsson, S. R., Simonsen, A. SNX18 tubulates recycling endosomes for autophagosome biogenesis. Autophagy. 9 (10), 1639-1641 (2013).
- Takahashi, Y., et al. The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget. 7 (15), 20855-20868 (2016).
- Imai, K., et al. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. Journal of Cell Science. 129 (20), 3781-3791 (2016).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- van Vliet, A. R., et al. ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation. Molecular Cell. 82 (22), 4324-4339 (2022).
- Kielkopf, C. L., Bauer, W., Urbatsch, I. L. Bradford assay for determining protein concentration. Cold Spring Harbor Protocols. 2020 (4), 102269 (2020).
- Kurien, B. T., Scofield, R. H. Validating antibody specificities for immunohistochemistry by protein blotting. Methods in Molecular Biology. 2593, 21-33 (2023).
- Lamb, C. A., et al. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO Journal. 35 (3), 281-301 (2016).
- Longatti, A., et al. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. Journal of Cell Biology. 197 (5), 659-675 (2012).
- Ravussin, A., Brech, A., Tooze, S. A., Stenmark, H. The phosphatidylinositol 3-phosphate-binding protein SNX4 controls ATG9A recycling and autophagy. Journal of Cell Science. 134 (3), (2021).
- DesMarais, V., Eddy, R. J., Sharma, V. P., Stone, O., Condeelis, J. S. Optimizing leading edge F-actin labeling using multiple actin probes, fixation methods and imaging modalities. BioTechniques. 66 (3), 113-119 (2019).
- Ennis, H. L., Lubin, M. Cycloheximide: Aspects of inhibition of protein synthesis in mammalian cells. Science. 146 (3650), 1474-1476 (1964).
- Webber, J. L., Tooze, S. A. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO Journal. 29 (1), 27-40 (2010).
- Claude-Taupin, A., et al. ATG9A protects the plasma membrane from programmed and incidental permeabilization. Nature Cell Biology. 23 (8), 846-858 (2021).
- Aaron, J. S., Taylor, A. B., Chew, T. L. Image co-localization - Co-occurrence versus correlation. Journal of Cell Science. 131 (3), 211847 (2018).
- D'Antuono, R., Nechyporuk-Zloy, V. Basic digital image acquisition, design, processing, analysis, management, and presentation. Principles of Light Microscopy: From Basic to Advanced. , 77-104 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved