A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Generating the Transcriptional Regulation View of Transcriptomic Features for Prediction Task and Dark Biomarker Detection on Small Datasets
In This Article
Summary
Here, we introduce a protocol for converting transcriptomic data into a mqTrans view, enabling the identification of dark biomarkers. While not differentially expressed in conventional transcriptomic analyses, these biomarkers exhibit differential expression in the mqTrans view. The approach serves as a complementary technique to traditional methods, unveiling previously overlooked biomarkers.
Abstract
Transcriptome represents the expression levels of many genes in a sample and has been widely used in biological research and clinical practice. Researchers usually focused on transcriptomic biomarkers with differential representations between a phenotype group and a control group of samples. This study presented a multitask graph-attention network (GAT) learning framework to learn the complex inter-genic interactions of the reference samples. A demonstrative reference model was pre-trained on the healthy samples (HealthModel), which could be directly used to generate the model-based quantitative transcriptional regulation (mqTrans) view of the independent test transcriptomes. The generated mqTrans view of transcriptomes was demonstrated by prediction tasks and dark biomarker detection. The coined term "dark biomarker" stemmed from its definition that a dark biomarker showed differential representation in the mqTrans view but no differential expression in its original expression level. A dark biomarker was always overlooked in traditional biomarker detection studies due to the absence of differential expression. The source code and the manual of the pipeline HealthModelPipe can be downloaded from http://www.healthinformaticslab.org/supp/resources.php.
Introduction
Transcriptome consists of the expressions of all the genes in a sample and may be profiled by high-throughput technologies like microarray and RNA-seq1. The expression levels of one gene in a dataset are called a transcriptomic feature, and the differential representation of a transcriptomic feature between the phenotype and control groups defines this gene as a biomarker of this phenotype2,3. Transcriptomic biomarkers have been extensively utilized in the investigations of disease diagnosis4, biological mechanism5, and survival analysis6,7, etc.
Gene activity patterns in the healthy tissues carry crucial information about the lives8,9. These patterns offer invaluable insights and act as ideal references for understanding the complex developmental trajectories of benign disorders10,11 and lethal diseases12. Genes interact with each other, and transcriptomes represent the final expression levels after their complicated interactions. Such patterns are formulated as transcriptional regulation network13 and metabolism network14, etc. The expressions of messenger RNAs (mRNAs) can be transcriptionally regulated by transcription factors (TFs) and long intergenic non-coding RNAs (lincRNAs)15,16,17. Conventional differential expression analysis ignored such complex gene interactions with the assumption of inter-feature independence18,19.
Recent advancements in graph neural networks (GNNs) demonstrate extraordinary potential in extracting important information from OMIC-based data for cancer studies20, e.g., identifying co-expression modules21. The innate capacity of GNNs renders them ideal for modeling the intricate relationships and dependencies among genes22,23.
Biomedical studies often focus on accurately predicting a phenotype against the control group. Such tasks are commonly formulated as binary classifications24,25,26. Here, the two class labels are typically encoded as 1 and 0, true and false, or even positive and negative27.
This study aimed to provide an easy-to-use protocol for generating the transcriptional regulation (mqTrans) view of a transcriptome dataset based on the pre-trained graph-attention network (GAT) reference model. The multitask GAT framework from a previously published work26 was used to transform transcriptomic features to the mqTrans features. A large dataset of healthy transcriptomes from the University of California, Santa Cruz (UCSC) Xena platform28 was used to pre-train the reference model (HealthModel), which quantitatively measured the transcription regulations from the regulatory factors (TFs and lincRNAs) to the target mRNAs. The generated mqTrans view could be used to build prediction models and detect dark biomarkers. This protocol utilizes the colon adenocarcinoma (COAD) patient dataset from The Cancer Genome Atlas (TCGA) database29 as an illustrative example. In this context, patients in stages I or II are categorized as negative samples, while those in stages III or IV are considered positive samples. The distributions of dark and traditional biomarkers across the 26 TCGA cancer types are also compared.
Description of the HealthModel pipeline
The methodology employed in this protocol is based on the previously published framework26, as outlined in Figure 1. To commence, users are required to prepare the input dataset, feed it into the proposed HealthModel pipeline, and obtain mqTrans features. Detailed data preparation instructions are provided in section 2 of the protocol section. Subsequently, users have the option to combine mqTrans features with the original transcriptomic features or proceed with the generated mqTrans features only. The produced dataset is then subjected to a feature selection process, with users having the flexibility to choose their preferred value for k in k-fold cross-validation for classification. The primary evaluation metric utilized in this protocol is accuracy.
HealthModel26 categorizes the transcriptomic features into three distinct groups: TF (Transcription Factor), lincRNA (long intergenic non-coding RNA), and mRNA (messenger RNA). The TF features are defined based on the annotations available in the Human Protein Atlas30,31. This work utilizes the annotations of lincRNAs from the GTEx dataset32. Genes belonging to the third-level pathways in the KEGG database33 are considered as mRNA features. It is worth noting that if an mRNA feature exhibits regulatory roles for a target gene as documented in the TRRUST database34, it is reclassified into the TF class.
This protocol also manually generates the two example files for the gene IDs of regulatory factors (regulatory_geneIDs.csv) and target mRNA (target_geneIDs.csv). The pairwise distance matrix among the regulatory features (TFs and lincRNAs) is calculated by the Pearson correlation coefficients and clustered by the popular tool weighted gene co-expression network analysis (WGCNA)36 (adjacent_matrix.csv). Users can directly utilize the HealthModel pipeline together with these example configuration files to generate the mqTrans view of a transcriptomic dataset.
Technical details of HealthModel
HealthModel represents the intricate relationships among TFs and lincRNAs as a graph, with the input features serving as the vertices denoted by V and an inter-vertex edge matrix designated as E. Each sample is characterized by K regulatory features, symbolized as VK×1. Specifically, the dataset encompassed 425 TFs and 375 lincRNAs, resulting in a sample dimensionality of K = 425 + 375 = 800. To establish the edge matrix E, this work employed the popular tool WGCNA35. The pairwise weight linking two vertices represented as 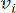 and
and 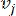 , is determined by the Pearson correlation coefficient. The gene regulatory network exhibits a scale-free topology36, characterized by the presence of hub genes with pivotal functional roles. We compute the correlation between two features or vertices,
, is determined by the Pearson correlation coefficient. The gene regulatory network exhibits a scale-free topology36, characterized by the presence of hub genes with pivotal functional roles. We compute the correlation between two features or vertices, 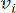 and
and 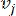 , using the topological overlap measure (TOM) as follows:
, using the topological overlap measure (TOM) as follows:
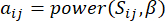 (1)
(1)
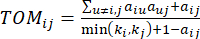 (2)
(2)
The soft threshold β is calculated using the 'pickSoft Threshold' function from the WGCNA package. The power exponential function aij is applied, where 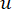 represents a gene excluding i and j, and
represents a gene excluding i and j, and 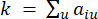 represents the vertex connectivity. WGCNA clusters the expression profiles of the transcriptomic features into multiple modules using a commonly employed dissimilarity measure (
represents the vertex connectivity. WGCNA clusters the expression profiles of the transcriptomic features into multiple modules using a commonly employed dissimilarity measure (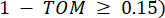 37.
37.
The HealthModel framework was originally designed as a multitask learning architecture26. This protocol only utilizes the model pre-training task for the construction of the transcriptomic mqTrans view. The user may choose to further refine the pre-trained HealthModel under the multitask graph attention network with additional task-specific transcriptomic samples.
Technical details of feature selection and classification
The feature selection pool implements eleven feature selection (FS) algorithms. Among them, three are filter-based FS algorithms: selecting K best features using the Maximal Information Coefficient (SK_mic), selecting K features based on the FPR of MIC (SK_fpr), and selecting K features with the highest false discovery rate of MIC (SK_fdr). Additionally, three tree-based FS algorithms assess individual features using a decision tree with the Gini index (DT_gini), adaptive boosted decision trees (AdaBoost), and random forest (RF_fs). The pool also incorporates two wrapper methods: Recursive feature elimination with the linear support vector classifier (RFE_SVC) and recursive feature elimination with the logistic regression classifier (RFE_LR). Finally, two embedding algorithms are included: linear SVC classifier with the top-ranked L1 feature importance values (lSVC_L1) and logistic regression classifier with the top-ranked L1 feature importance values (LR_L1).
The classifier pool employs seven different classifiers to build classification models. These classifiers comprise linear support vector machine (SVC), Gaussian Naïve Bayes (GNB), logistic regression classifier (LR), k-nearest neighbor, with k set to 5 by default (KNN), XGBoost, random forest (RF), and decision tree (DT).
The random split of the dataset into the train: test subsets can be set in the command line. The demonstrated example uses the ratio of train: test = 8: 2.
Access restricted. Please log in or start a trial to view this content.
Protocol
NOTE: The following protocol describes the details of the informatics analytic procedure and Python commands of the major modules. Figure 2 illustrates the three major steps with example commands utilized in this protocol and refer to previously published works26,38 for more technical details. Do the following protocol under a normal user account in a computer system and avoid using the administrator or root account. This is a computational protocol and has no biomedical hazardous factors.
1. Prepare Python environment
- Create a virtual environment.
- This study used the Python programming language and a Python virtual environment (VE) with Python 3.7. Follow these steps (Figure 3A):
conda create -n healthmodel python=3.7
conda create is the command to create a new VE. The parameter -n specifies the name of the new environment, in this case, healthmodel. And python=3.7 specifies the Python version to be installed. Choose any preferred name and Python version supporting the above command. - After running the command, the output is similar to Figure 3B. Enter y and wait for the process to complete.
- This study used the Python programming language and a Python virtual environment (VE) with Python 3.7. Follow these steps (Figure 3A):
- Activate the virtual environment
- In most cases, activate the created VE with the following command (Figure 3C):
conda activate healthmodel - Follow the platform-specific instructions for the VE activation, if some platforms require the user to upload the platform-specific configuration files for activation.
- In most cases, activate the created VE with the following command (Figure 3C):
- Install PyTorch 1.13.1
- PyTorch is a popular Python package for artificial intelligence (AI) algorithms. Use PyTorch 1.13.1, based on the CUDA 11.7 GPU programming platform, as an example. Find other versions at https://pytorch.org/get-started/previous-versions/. Use the following command (Figure 3D):
pip3 install torch torchvision torchaudio
NOTE: Using PyTorch version 1.12 or newer is strongly recommended. Otherwise, installing the required package torch_geometric may be challenging, as noted on the official torch_geometric website: https://pytorch-geometric.readthedocs.io/en/latest/install/installation.html.
- PyTorch is a popular Python package for artificial intelligence (AI) algorithms. Use PyTorch 1.13.1, based on the CUDA 11.7 GPU programming platform, as an example. Find other versions at https://pytorch.org/get-started/previous-versions/. Use the following command (Figure 3D):
- Install additional packages for torch-geometric
- Following the guidelines at https://pytorch-geometric.readthedocs.io/en/latest/install/installation.html, install the following packages: torch_scatter, torch_sparse, torch_cluster, and torch_spline_conv using the command (Figure 3E):
pip install pyg_lib torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-1.13.0+cu117.html
- Following the guidelines at https://pytorch-geometric.readthedocs.io/en/latest/install/installation.html, install the following packages: torch_scatter, torch_sparse, torch_cluster, and torch_spline_conv using the command (Figure 3E):
- Install torch-geometric package.
- This study requires a specific version, 2.2.0, of the torch-geometric package. Run the command (Figure 3F):
pip install torch_geometric==2.2.0
- This study requires a specific version, 2.2.0, of the torch-geometric package. Run the command (Figure 3F):
- Install other packages.
- Packages like pandas are usually available by default. If not, install them using the pip command. For instance, to install pandas and xgboost, run:
pip install pandas
pip install xgboost
- Packages like pandas are usually available by default. If not, install them using the pip command. For instance, to install pandas and xgboost, run:
2. Using the pre-trained HealthModel to generate the mqTrans features
- Download the code and the pre-trained model.
- Download the code and the pre-trained HealthModel from the website: http://www.healthinformaticslab.org/supp/resources.php, which is named HealthModel-mqTrans-v1-00.tar.gz (Figure 4A). The downloaded file can be decompressed to a user-specified path. The detailed formulation and the supporting data of the implemented protocol can be found in26.
- Introduce the parameters to run HealthModel.
- Firstly, change the working directory to the HealthModel-mqTrans folder in the command line. Use the following syntax for running the code:
python main.py <data folder> <model folder> <output folder>
The details regarding each parameter and the data, model and output folders are as follows:
data folder: This is the source data folder, and each data file is in the csv format. This data folder has two files (see detailed descriptions in steps 2.3 and 2.4). These files need to be replaced with personal data.
data.csv: The transcriptomic matrix file. The first row lists the feature (or gene) IDs, and the first column gives the sample IDs. The list of genes includes the regulatory factors (TFs and lincRNAs), and the regulated mRNA genes.
label.csv: The sample label file. The first column lists the sample IDs, and the column with the name "label" gives the sample label.
model folder: The folder to save information about the model:
HealthModel.pth: The pre-trained HealthModel.
regulatory_geneIDs.csv: The regulatory gene IDs used in this study.
target_geneIDs.csv: The target genes used in this study.
adjacent_matrix.csv: The adjacent matrix of regulatory genes.
output folder: The output files are written to this folder, created by the code.
test_target.csv: The gene expression value of target genes after Z-Normalization and imputation.
pred_target.csv: The predicted gene expression value of target genes.
mq_target.csv: The predicted gene expression value of target genes.
- Firstly, change the working directory to the HealthModel-mqTrans folder in the command line. Use the following syntax for running the code:
- Prepare the transcriptomic matrix file in the csv format.
- Each row represents a sample, and each column represents a gene (Figure 4B). Name the transcriptomic data matrix file as data.csv in the data folder.
NOTE: This file may be generated by manually saving a data matrix in the .csv format from software such as Microsoft Excel. The transcriptomic matrix may also be generated by computer programming.
- Each row represents a sample, and each column represents a gene (Figure 4B). Name the transcriptomic data matrix file as data.csv in the data folder.
- Prepare the label file in the csv format.
- Similar to the transcriptomic matrix file, name the label file as label.csv in the data folder (Figure 4C).
NOTE: The first column gives the sample names, and the class label of each sample is given in the column titled label. The 0 value in the label column means this sample is negative, 1 means a positive sample.
- Similar to the transcriptomic matrix file, name the label file as label.csv in the data folder (Figure 4C).
- Generate the mqTrans features.
- Run the following command to generate the mqTrans features and get the outputs shown in Figure 4D. The mqTrans features are generated as the file ./output/mq_targets.csv, and the label file is resaved as the file ./output/label.csv. For the convenience of further analysis, the original expression values of the mRNA genes are also extracted as the file ./output/ test_target.csv.
python ./Get_mqTrans/code/main.py ./data ./Get_mqTrans/model ./output
- Run the following command to generate the mqTrans features and get the outputs shown in Figure 4D. The mqTrans features are generated as the file ./output/mq_targets.csv, and the label file is resaved as the file ./output/label.csv. For the convenience of further analysis, the original expression values of the mRNA genes are also extracted as the file ./output/ test_target.csv.
3. Select mqTrans Features
- Syntax of the feature selection code
- Firstly, change the working directory to the HealthModel-mqTrans folder. Use the following syntax:
python ./FS_classification/testMain.py <in-data-file.csv> <in-label-file.csv> <output folder> <select_feature_number> <test_size> <combine> <combine file>
The details of each parameter are as follows:
in-data-file: The input data file
in-label-file: The label of the input data file
output folder: Two output files are saved in this folder, including Output-score.xlsx (the feature selection method and the accuracy of the corresponding classifier), and Output-SelectedFeatures.xlsx (the selected feature names for each feature selection algorithm).- select_feature_number: select the number of features, ranging from 1 to the number of the features of the data file.
- test_size: Set the ratio of the test sample to split. For example, 0.2 means that the input dataset is randomly split into the train: test subsets by the ratio of 0.8:0.2.
- combine: If true, combine two data files together for feature selection, i.e., the original expression values and the mqTrans features. If false, just use one data file for feature selection, i.e., the original expression values or the mqTrans features.
- combine file: If combine is true, provide this file name to save the combined data matrix.
NOTE: This pipeline aims to demonstrate how the generated mqTrans features perform on classification tasks, and it directly uses the file generated by section 2 for the following operations.
- Firstly, change the working directory to the HealthModel-mqTrans folder. Use the following syntax:
- Run feature selection algorithm for mqTrans feature selection.
- Turn combine =False if the user selects mqTrans features or original features.
- Firstly, select 800 original features and split the dataset into train: test=0.8:0.2:
python ./FS_classification/testMain.py ./output/test_target.csv ./output/label.csv ./result 800 0.2 False - Turn combine =True, if the user wants to combine the mqTrans features with the original expression values to select features. Here, the demonstrative example is to select 800 features and split the dataset into train: test=0.8:0.2:
python ./FS_classification/testMain.py ./output/mq_targets.csv ./output/label.csv ./result_combine 800 0.2 True ./output/test_target.csv
NOTE: Figure 5 shows the output information. The supplementary files required for this protocol are in HealthModel-mqTrans-v1-00.tar folder (Supplementary Coding File 1).
Access restricted. Please log in or start a trial to view this content.
Results
Evaluation of the mqTrans view of the transcriptomic dataset
The test code uses eleven feature selection (FS) algorithms and seven classifiers to evaluate how the generated mqTrans view of the transcriptomic dataset contributes to the classification task (Figure 6). The test dataset consists of 317 colon adenocarcinoma (COAD) from The Cancer Genome Atlas (TCGA) database29. The COAD patients at stages I or II are regarded as the negative samples,...
Access restricted. Please log in or start a trial to view this content.
Discussion
Section 2 (Use the pre-trained HealthModel to generate the mqTrans features) of the protocol is the most critical step within this protocol. After preparing the computational working environment in section 1, section 2 generates the mqTrans view of a transcriptomic dataset based on the pre-trained large reference model. Section 3 is a demonstrative example of selecting the generated mqTrans features for biomarker detections and prediction tasks. The users can conduct other transcriptomic analyses on this mqTrans dataset ...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Senior and Junior Technological Innovation Team (20210509055RQ), Guizhou Provincial Science and Technology Projects (ZK2023-297), the Science and Technology Foundation of Health Commission of Guizhou Province (gzwkj2023-565), Science and Technology Project of Education Department of Jilin Province (JJKH20220245KJ and JJKH20220226SK), the National Natural Science Foundation of China (U19A2061), the Jilin Provincial Key Laboratory of Big Data Intelligent Computing (20180622002JC), and the Fundamental Research Funds for the Central Universities, JLU. We extend our sincerest appreciation to the review editor and the three anonymous reviewers for their constructive critiques, which have been instrumental in substantially enhancing the rigor and clarity of this protocol.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Anaconda | Anaconda | version 2020.11 | Python programming platform |
| Computer | N/A | N/A | Any general-purpose computers satisfy the requirement |
| GPU card | N/A | N/A | Any general-purpose GPU cards with the CUDA computing library |
| pytorch | Pytorch | version 1.13.1 | Software |
| torch-geometric | Pytorch | version 2.2.0 | Software |
References
- Mutz, K. -O., Heilkenbrinker, A., Lönne, M., Walter, J. -G., Stahl, F. Transcriptome analysis using next-generation sequencing. Curr Opin in Biotechnol. 24 (1), 22-30 (2013).
- Meng, G., Tang, W., Huang, E., Li, Z., Feng, H. A comprehensive assessment of cell type-specific differential expression methods in bulk data. Brief Bioinform. 24 (1), 516(2023).
- Iqbal, N., Kumar, P. Integrated COVID-19 Predictor: Differential expression analysis to reveal potential biomarkers and prediction of coronavirus using RNA-Seq profile data. Comput Biol Med. 147, 105684(2022).
- Ravichandran, S., et al. VB(10), a new blood biomarker for differential diagnosis and recovery monitoring of acute viral and bacterial infections. EBioMedicine. 67, 103352(2021).
- Lv, J., et al. Targeting FABP4 in elderly mice rejuvenates liver metabolism and ameliorates aging-associated metabolic disorders. Metabolism. 142, 155528(2023).
- Cruz, J. A., Wishart, D. S. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2, 59-77 (2007).
- Cox, D. R. Analysis of Survival Data. , Chapman and Hall/CRC. London. (2018).
- Newman, A. M., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 12 (5), 453-457 (2015).
- Ramilowski, J. A., et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 6 (1), 7866(2015).
- Xu, Y., et al. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate. 77 (10), 1167-1175 (2017).
- Wang, Y., et al. Profiles of differential expression of circulating microRNAs in hepatitis B virus-positive small hepatocellular carcinoma. Cancer Biomark. 15 (2), 171-180 (2015).
- Hu, S., et al. Transcriptional response profiles of paired tumor-normal samples offer novel. Oncotarget. 8 (25), 41334-41347 (2017).
- Xu, H., Luo, D., Zhang, F. DcWRKY75 promotes ethylene induced petal senescence in carnation (Dianthus caryophyllus L). Plant J. 108 (5), 1473-1492 (2021).
- Niu, H., et al. Dynamic role of Scd1 gene during mouse oocyte growth and maturation. Int J Biol Macromol. 247, 125307(2023).
- Aznaourova, M., et al. Single-cell RNA sequencing uncovers the nuclear decoy lincRNA PIRAT as a regulator of systemic monocyte immunity during COVID-19. Proc Natl Acad Sci U S A. 119 (36), 2120680119(2022).
- Prakash, A., Banerjee, M. An interpretable block-attention network for identifying regulatory feature interactions. Brief Bioinform. 24 (4), (2023).
- Zhai, Y., et al. Single-cell RNA sequencing integrated with bulk RNA sequencing analysis reveals diagnostic and prognostic signatures and immunoinfiltration in gastric cancer. Comput Biol Med. 163, 107239(2023).
- Duan, L., et al. Dynamic changes in spatiotemporal transcriptome reveal maternal immune dysregulation of autism spectrum disorder. Comput Biol Med. 151, 106334(2022).
- Zolotareva, O., et al. Flimma: a federated and privacy-aware tool for differential gene expression analysis). Genome Biol. 22 (1), 338(2021).
- Su, R., Zhu, Y., Zou, Q., Wei, L. Distant metastasis identification based on optimized graph representation of gene. Brief Bioinform. 23 (1), (2022).
- Xing, X., et al. Multi-level attention graph neural network based on co-expression gene modules for disease diagnosis and prognosis. Bioinformatics. 38 (8), 2178-2186 (2022).
- Bongini, P., Pancino, N., Scarselli, F., Bianchini, M. BioGNN: How Graph Neural Networks Can Solve Biological Problems. Artificial Intelligence and Machine Learning for Healthcare: Vol. 1: Image and Data Analytics. , Springer. Cham. (2022).
- Muzio, G., O'Bray, L., Borgwardt, K. Biological network analysis with deep learning. Brief Bioinform. 22 (2), 1515-1530 (2021).
- Luo, H., et al. Multi-omics integration for disease prediction via multi-level graph attention network and adaptive fusion. bioRxiv. , (2023).
- Feng, X., et al. Selecting multiple biomarker subsets with similarly effective binary classification performances. J Vis Exp. (140), e57738(2018).
- Duan, M., et al. Orchestrating information across tissues via a novel multitask GAT framework to improve quantitative gene regulation relation modeling for survival analysis. Brief Bioinform. 24 (4), (2023).
- Chicco, D., Starovoitov, V., Jurman, G. The benefits of the Matthews correlation Coefficient (MCC) over the diagnostic odds ratio (DOR) in binary classification assessment. IEEE Access. 9, 47112-47124 (2021).
- Goldman, M. J., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 38 (6), 675-678 (2020).
- Liu, J., et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 173 (2), 400-416 (2018).
- Uhlen, M., et al. Towards a knowledge-based human protein atlas. Nat Biotechnol. 28 (12), 1248-1250 (2010).
- Hernaez, M., Blatti, C., Gevaert, O. Comparison of single and module-based methods for modeling gene regulatory. Bioinformatics. 36 (2), 558-567 (2020).
- Consortium, G. The genotype-tissue expression (GTEx) project. Nat Genet. 45 (6), 580-585 (2013).
- Kanehisa, M., et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587-D592 (2023).
- Han, H., et al. TRRUST v2: an expanded reference database of human and mouse transcriptional. Nucleic Acids Res. 46, D380-D386 (2018).
- Langfelder, P., Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9, 559(2008).
- Sulaimanov, N., et al. Inferring gene expression networks with hubs using a degree weighted Lasso. Bioinformatics. 35 (6), 987-994 (2019).
- Kogelman, L. J. A., Kadarmideen, H. N. Weighted Interaction SNP Hub (WISH) network method for building genetic networks. BMC Syst Biol. 8, 5(2014).
- Duan, M., et al. Pan-cancer identification of the relationship of metabolism-related differentially expressed transcription regulation with non-differentially expressed target genes via a gated recurrent unit network. Comput Biol Med. 148, 105883(2022).
- Duan, M., et al. Detection and independent validation of model-based quantitative transcriptional regulation relationships altered in lung cancers. Front Bioeng Biotechnol. 8, 582(2020).
- Fiorini, N., Lipman, D. J., Lu, Z. Towards PubMed 2.0. eLife. 6, 28801(2017).
- Liu, J., et al. Maternal microbiome regulation prevents early allergic airway diseases in mouse offspring. Pediatr Allergy Immunol. 31 (8), 962-973 (2020).
- Childs, E. J., et al. Association of common susceptibility variants of pancreatic cancer in higher-risk patients: A PACGENE study. Cancer Epidemiol Biomarkers Prev. 25 (7), 1185-1191 (2016).
- Wang, C., et al. Thailandepsins: bacterial products with potent histone deacetylase inhibitory activities and broad-spectrum antiproliferative activities. J Nat Prod. 74 (10), 2031-2038 (2011).
- Lv, X., et al. Transcriptional dysregulations of seven non-differentially expressed genes as biomarkers of metastatic colon cancer. Genes (Basel). 14 (6), 1138(2023).
- Li, X., et al. Undifferentially expressed CXXC5 as a transcriptionally regulatory biomarker of breast cancer. Advanced Biology. , (2023).
- Yuan, W., et al. The N6-methyladenosine reader protein YTHDC2 promotes gastric cancer progression via enhancing YAP mRNA translation. Transl Oncol. 16, 101308(2022).
- Tanabe, A., et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 376 (1), 34-42 (2016).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved