Se requiere una suscripción a JoVE para ver este contenido. Inicie sesión o comience su prueba gratuita.
Method Article
Medición de la longitud de la cola de Poly A a partir del cerebro y la línea celular de la larva de Drosophila
En este artículo
Resumen
El protocolo describe un método eficiente y fiable para cuantificar la longitud poli(A) del gen de interés del sistema nervioso de Drosophila , que puede adaptarse fácilmente a tejidos o tipos celulares de otras especies.
Resumen
La poliadenilación es una modificación postranscripcional crucial que agrega colas de poli(A) al extremo 3' de las moléculas de ARNm. La longitud de la cola de poli(A) está estrechamente regulada por procesos celulares. La desregulación de la poliadenilación del ARNm se ha asociado con una expresión génica anormal y diversas enfermedades, como el cáncer, los trastornos neurológicos y las anomalías del desarrollo. Por lo tanto, comprender la dinámica de la poliadenilación es vital para desentrañar las complejidades del procesamiento del ARNm y la regulación génica postranscripcional.
Este artículo presenta un método para medir la longitud de la cola de poli(A) en muestras de ARN aisladas de cerebros de larvas de Drosophila y células S2 de Drosophila Schneider. Empleamos el enfoque de relavesa guanosina/inosina (G/I), que implica la adición enzimática de residuos G/I en el extremo 3' del ARNm utilizando poli(A) polimerasa de levadura. Esta modificación protege el extremo 3' del ARN de la degradación enzimática. A continuación, las colas de polietileno (A) protegidas se transcriben a la inversa utilizando un cebador antisentido universal. Posteriormente, la amplificación por PCR se realiza utilizando un oligonucleótido específico del gen que se dirige al gen de interés, junto con un oligonucleótido de secuencia universal utilizado para la transcripción inversa.
Esto genera productos de PCR que abarcan las colas de poli(A) del gen de interés. Dado que la poliadenilación no es una modificación uniforme y da lugar a colas de diferentes longitudes, los productos de PCR muestran una gama de tamaños, lo que da lugar a un patrón de frotis en el gel de agarosa. Por último, los productos de PCR se someten a electroforesis en gel capilar de alta resolución, seguida de la cuantificación utilizando los tamaños de los productos de PCR poli(A) y el producto de PCR específico del gen. Esta técnica ofrece una herramienta sencilla y fiable para analizar las longitudes de la cola de poli(A), lo que nos permite obtener una visión más profunda de los intrincados mecanismos que gobiernan la regulación del ARNm.
Introducción
La mayoría de los ARNm eucariotas están poliadenilados postranscripcionalmente en su extremo 3' en el núcleo mediante la adición de adenosinas no moldeadas por poli(A) polimerasas canónicas. Una cola de poli(A) intacta es fundamental a lo largo del ciclo de vida del ARNm, ya que es esencial para la exportación nuclear de ARNm1, facilita la interacción con las proteínas de unión a poli(A) para mejorar la eficiencia de la traducción2 e imparte resistencia contra la degradación3. En ciertos casos, la cola de poli(A) también puede sufrir una extensión en el citoplasma, facilitada por poli(A) polimerasas no canónicas4. En el citoplasma, la longitud de la cola de poli (A) cambia dinámicamente e influye en la vida útil de la molécula de ARNm. Numerosas polimerasas y deadenilasas son conocidas por modular la longitud de la cola 5,6,7. Por ejemplo, el acortamiento de las colas de poli(A) se correlaciona con la represión de la traducción, mientras que el alargamiento de las colas de poli(A) mejora la traslación 8,9.
Los estudios genómicos acumulados han demostrado la importancia fundamental de la longitud de la cola de poli(A) en varias facetas de la biología eucariota. Esto incluye el papel en el desarrollo de las células germinales, el desarrollo embrionario temprano, la plasticidad sináptica neuronal para el aprendizaje y la memoria, y la respuesta inflamatoria10. Se han desarrollado numerosos métodos y ensayos para medir las longitudes de la cola de poli(A). Por ejemplo, el ensayo RNasa H/oligo(dT) aprovecha la RNasa H en presencia o ausencia de oligo(dT) para estudiar la longitud de la cola de poli(A) 11,12. Otros métodos para estudiar la cola de poli(A) incluyen la amplificación por PCR de los extremos 3', como la prueba de amplificación rápida de los extremos del ADNc (RACE-PAT)12,13 y la prueba de poli(A) mediada por ligasa (LM-PAT)14. Otras modificaciones del ensayo PAT incluyen ePAT15 y sPAT16. El G-tailingenzimático 17,18 o el G/I tailing del extremo 3' son otras variaciones del ensayo PAT. La modificación adicional de estas técnicas incluye el uso de cebadores marcados con fluorescencia junto con electroforesis en gel capilar para el análisis de alta resolución, lo que se conoce como prueba de poli(A) de alta resolución (Hire-PAT)19. Estos ensayos impulsados por PCR permiten una cuantificación rápida y de alta sensibilidad de la longitud del poli(A).
Con el desarrollo de la secuenciación de próxima generación, un método de secuenciación de alto rendimiento, como PAL-seq20 y TAIL-seq21, permite realizar análisis de poliadenilación a escala de todo el transcriptoma. Sin embargo, estos métodos solo proporcionan lecturas cortas de secuenciación de 36-51 nucleótidos. Por lo tanto, FLAM-Seq22 se desarrolló para el perfil global de la longitud de la cola del ARNm de longitud completa y proporciona lecturas largas. La tecnología Nanopore23 proporciona secuenciación directa de ARN o ADNc independiente de la PCR para estimaciones de la longitud de la cola poli(A). Sin embargo, estos métodos de alto rendimiento no están exentos de limitaciones. Requieren grandes cantidades de materiales de partida, son caros y requieren mucho tiempo. Además, el análisis de transcripciones raras puede ser extremadamente difícil con los métodos de alto rendimiento, y los métodos basados en PCR de bajo rendimiento siguen proporcionando una ventaja cuando es necesario analizar un pequeño número de transcripciones, para experimentos piloto y validación de otros métodos.
Recientemente hemos demostrado que los ARNm de Dscam1 contienen colas cortas de poli(A) en Drosophila, lo que requiere una unión no canónica de la proteína de unión a poli(A) citoplasmática en Dscam1 3'UTR utilizando el método de relaves G/I24. Aquí proporcionamos un procedimiento simplificado para la preparación de tejidos y la cuantificación de la longitud poli(A) de los ARNm del sistema nervioso de Drosophila y las células S2 de Drosophila .
Access restricted. Please log in or start a trial to view this content.
Protocolo
1. Cría y selección de larvas de Drosophila
- Mantener/cultivar la cepa de mosca (w1118, wildtype) en un medio estándar de alimentación para moscas a 25 °C en una incubadora humidificada.
- Seleccione 10 larvas errantes de 3er estadio 72 h después de la puesta de huevos.
- Coloque las larvas en una placa de Petri vacía de 35 mm y lávelas suavemente transfiriendo las larvas a la nueva placa que contiene agua del grifo con fórceps. Haga esto 2 veces para eliminar cualquier resto de comida.
2. Aislamiento cerebral de larvas de Drosophila (Figura 1)
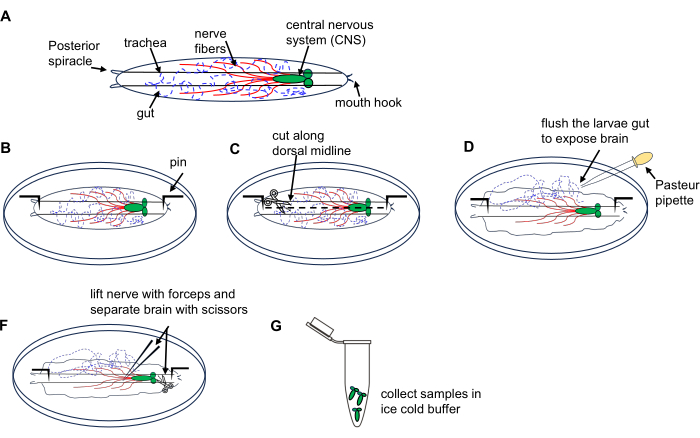
Figura 1: Disección del cerebro de la larva de Drosophila de la etapa de deambulación del3er estadio. (A) Dibujos esquemáticos de la larva de Drosophila. (B-G) Disección de larvas. Haga clic aquí para ver una versión más grande de esta figura.
- Coloque 10 larvas en un plato de disección que contenga PBS helado.
- Coloque la larva con el lado dorsal hacia arriba (identificado por los tubos traqueales que corren a lo largo de su longitud) y sujete cada extremo al fondo del plato, seguido de hacer una pequeña incisión en la pared del cuerpo en el extremo posterior.
- Corta la pared del cuerpo a lo largo de la línea media dorsal hacia el extremo anterior con unas tijeras de microdisección.
- Enjuague brevemente el interior de la larva con una pipeta Pasteur 3x con PBS en el plato para exponer el cerebro.
- Localice y levante el cerebro con las pinzas y aíslelo cuidadosamente con unas tijeras de microdisección.
- Transfiera los cerebros disecados a un tubo de microcentrífuga de 1,5 ml lleno de PBS helado en hielo; Recoge todos los cerebros de larvas. Proceda con la extracción de ARN utilizando un kit de micropreparación de ARN como se describe en la sección 4.
NOTA: Realice una disección de 10 larvas en 15 minutos para evitar daños en los tejidos y la degradación del ARN.
3. Células de Drosophila S2 Schneider
- Cultivar células de Drosophila S2 en medio de Drosophila Schneider suplementado con suero fetal bovino (FBS) al 10% a 25 °C en una incubadora humidificada hasta una densidad de 8 × 106 a 10 × 106 células/ml con un mínimo de 90% de viabilidad.
- En un tubo cónico estéril de 50 ml, diluir las células a 2,5 × 106 células/ml con medio Drosophila de Schneider suplementado con FBS al 10% que se ha precalentado a 25 °C.
- Transfiera 8 mL de la suspensión celular (20 × 106 células) a una placa de cultivo de 100 mm y agregue 4 mL de medio para llegar a 12 mL (día 1).
- Incubar las células cultivadas a 25 °C en una incubadora humidificada.
NOTA: Las células se adhieren libremente a la placa después de 12-16 h (día 2). - Transfectar las células con plásmidos de ADN apropiados24.
- Incubar durante 48 h en una incubadora humidificada.
- Después de la incubación, recoja las células añadiendo 5 ml de PBS helado mediante un pipeteo suave (día 4).
- Transfiera las células a un tubo de 15 ml.
- Granular las células centrifugando a 1.000 × g durante 5 min a 4 °C.
- Enjuague las células 2 veces con PBS helado mediante un pipeteo suave y recoja las células centrifugando a 1.000 × g durante 5 min a 4 °C.
- Realice la extracción de ARN con un kit de minipreparación de ARN.
NOTA: Realice los siguientes pasos dentro de una campana de flujo laminar estéril.
4. Extracción de ARN total del cerebro de las larvas de Drosophila y de las células S2
- Cerebro larvario: Eliminar el PBS mediante centrifugación breve (centrifugado corto de 8 s a 5.000 × g).
- Añadir 600 μL de tampón de lisis de ARN y homogeneizar 10x con un mortero de plástico. Inspeccione visualmente el tubo bajo un microscopio estereoscópico para garantizar una lisis completa.
- Centrifugar a 1.000 × g durante 5 min a 4 °C para eliminar los restos de tejido. Transfiera el sobrenadante aclarado a un tubo de microcentrífuga libre de nucleasa.
- Aísle el ARN con un kit de micropreparación de ARN de acuerdo con las instrucciones del fabricante.
NOTA: El uso de un kit de micropreparación de ARN es esencial para las muestras de cerebro larvario debido a la pequeña cantidad de ARN presente en las muestras. - Celdas S2: Retire el PBS y aísle el ARN de acuerdo con las instrucciones del fabricante.
- Mida el rendimiento y la calidad del ARN mediante espectrofotometría y electroforesis en gel de agarosa.
- Determine la pureza y la cantidad de ARN extraído midiendo la densidad óptica del ARN extraído a A260 nm y A280 nm, respectivamente. Asegúrese de que la relación A260 nm/A280 nm sea ≥2,0 y que la concentración de ARN sea de >350 ng/μL para aplicaciones posteriores.
NOTA: Un rendimiento típico de ARN de 10 cerebros de larvas de Drosophila es de ~500-800 ng/μL o 2,5-4 μg en 5 μL. Para las células S2, el rendimiento es de ~2-3 μg/μL (15-30 μg en 15 μL). El ARN aislado puede almacenarse a -80 °C para su almacenamiento a largo plazo.
- Determine la pureza y la cantidad de ARN extraído midiendo la densidad óptica del ARN extraído a A260 nm y A280 nm, respectivamente. Asegúrese de que la relación A260 nm/A280 nm sea ≥2,0 y que la concentración de ARN sea de >350 ng/μL para aplicaciones posteriores.
5. Preparación del gel de ARN y electroforesis
- Gel de ARN desnaturalizante al 1,5% (100 ml)
NOTA: El formaldehído es tóxico a través del contacto con la piel y la inhalación de vapores; Manéjelo en una campana de extracción química.- Disuelva tres comprimidos de agarosa (1,5 g) en 82 ml de tampón MOPS (Archivo suplementario 1) hasta que los comprimidos se rompan por completo para formar partículas finas.
- Calienta la suspensión de agarosa en un microondas hasta que la solución esté clara y todas las partículas se disuelvan por completo.
- Enfriar la solución a ~60 °C.
- Agregue 18 ml de formaldehído al 37%, luego mezcle agitando suavemente. Vierta la solución en la bandeja de fundición y deje que se solidifique en una campana extractora.
- Preparación de muestras de ARN y electroforesis
- Diluir la muestra de ARN a 200 ng (en 5 μL) y añadir 5 μL de colorante de carga de ARN 2x.
- Calentar las muestras a 70 °C durante 5 min en un baño seco.
- Cargue 2 μL de escalera de ARN en el primer carril y 10 μL de muestras en los carriles adyacentes.
- Realizar electroforesis en tampón MOPS a 100 V durante 60 min para un gel de 5 x 6 cm.
NOTA: Ajuste las condiciones de electroforesis en función de los tamaños de los amplicones. - Visualiza el gel en un transiluminador UV.
NOTA: La presencia de un tamaño de nucleótido de ~600 bandas individuales indica una preparación de ARN intacta (consulte la Figura 2A).
6. Medición de la longitud de la cola de Poly (A)
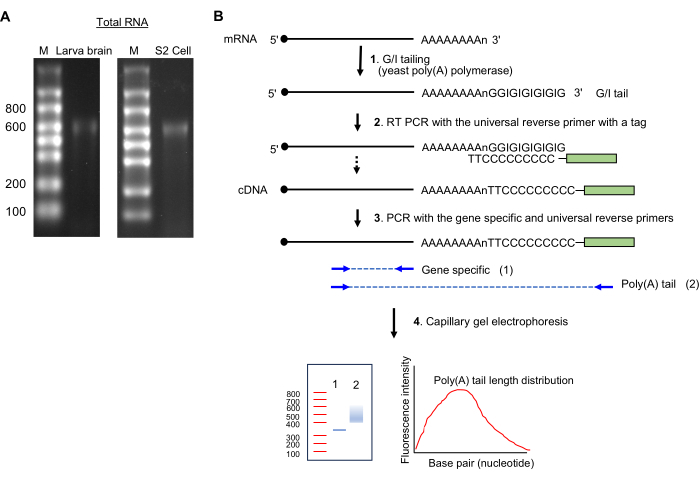
Figura 2: Preparación de la muestra de ARN y ensayo de cola de poli(A). (A) Las imágenes de gel de ARN muestran el ARN total del cerebro de la larva de Drosophila (izquierda) y las células S2 (derecha) en un gel de agarosa con formaldehído al 1,5%. Los tamaños de la escalera de ARN monocatenario se muestran en nucleótidos en el carril M. Obsérvese una banda de ARN importante a ~600 nt, que proviene del ARNr. (B) Esquemas del ensayo de cola de poli(A). Abreviatura: G/I = guanosina/inosina. Haga clic aquí para ver una versión más grande de esta figura.
- Relaves GI (Figura 2B)
- Manteniendo los reactivos en hielo, prepare la siguiente mezcla (20 μL): hasta 14 μL de muestra total de ARN (1 μg), 4 μL de 5x Mezcla de tampón de cola y 2 μL de mezcla de enzimas de cola 10x.
- Incubar a 37 °C durante 60 min en un termociclador.
- Añadir 1,5 μL de la solución de tope de cola; Mantener en hielo durante 2 min.
NOTA: Proceda a la transcripción inversa o almacene las muestras de ARN con cola GI a -80 °C hasta que estén listas para proceder a la transcripción inversa.
- Transcripción inversa y amplificación por PCR
- Sintetizar ADNc preparando la mezcla e incubándola en las condiciones descritas en el Archivo Suplementario 1.
- Diluir las muestras de ADNc y realizar la PCR para amplificar el ADN como se indica en el Archivo Suplementario 1.
7. Análisis del producto de PCR mediante electroforesis en gel de agarosa
- Analice una pequeña porción (2-5 μL) de los productos de PCR del paso 6.2.2 en un gel de agarosa al 2,5% mediante electroforesis a 100 V durante 45 min para el control de calidad.
- Verificar la especificidad de la PCR para reacciones específicas de genes y de cola mediante la secuenciación de las bandas de PCR extraídas en gel.
8. Electroforesis capilar
- Realice electroforesis en gel de alta resolución en 1 μl de productos de PCR (0,5-5 ng/μl) a partir de PCR específica de genes y poli(A) utilizando el bioanalizador con un kit de ADN de alta sensibilidad. Busque picos bien resueltos que indiquen una ejecución exitosa.
9. Análisis de datos: medición de la longitud de la cola poli(A) (Figura 3)
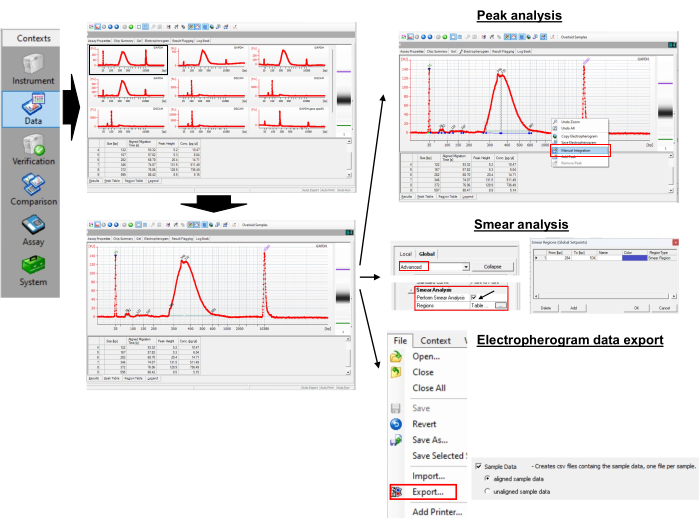
Figura 3: Medición de la longitud de la cola de Poly(A) y del valor máximo. Haga clic aquí para ver una versión más grande de esta figura.
- Acceso a los datos
- Para acceder a los datos, abra el archivo xad en el software.
- Seleccione el nombre de la muestra o la escalera en el panel de vista de árbol.
NOTA: Esto mostrará el resultado como electroferogramas o imágenes gelatinosas para las muestras seleccionadas. El marcador inferior 35 par de bases (pb) y el marcador superior 10.380 pb son los estándares internos utilizados para alinear los datos de la escalera (50-7.000 pb) con los datos de los pocillos de muestra. - Acérquese y aléjese de los electroferogramas y las imágenes gelatinosas para mostrar los detalles.
- Realización de análisis de picos y frotis
- Para obtener los tamaños de pico, abra el electroferograma de una muestra seleccionada.
- Haga clic con el botón derecho en el electroferograma y seleccione integración manual para seleccionar manualmente los picos arrastrando la línea horizontal.
- Observe los valores máximos de la tabla Picos. Identifique el pico con la mayor altura de pico. Este es un pico de longitud de cola de poli(A) para una muestra individual. En el ejemplo mostrado, es de 346 pb.
- Habilite el icono Mostrar/Ocultar puntos de ajuste en el menú superior y espere a que aparezca un nuevo panel en el lado derecho.
- Seleccione Avanzado, desplácese hacia abajo hasta encontrar Realizar análisis de frotis y active la casilla de verificación. Esto agregará la tabla Región a la pestaña de electroferograma.
- Seleccione un electroferograma de una muestra y vaya a la tabla Región, que muestra el menú Desde [bp] y Hasta [bp]. Para establecer el inicio y el final [bp], haga clic con el botón derecho en el electroferograma y seleccione Región para agregar Agregar región.
- Haga clic con el botón derecho en cualquier celda de la tabla Región y seleccione Modificar regiones para que aparezca una pequeña ventana nueva donde se pueden establecer regiones personalizadas.
NOTA: Por ejemplo, utilizamos una región de 300 pb a 550 pb para GAPDH. La PCR GAPDH específica del gen arrojó un pico de 265 pb. El cebador universal (Tabla 1) extiende la longitud de la PCR poli(A) en 35 pb a través del recocido a los ARN de cola G/I. Por lo tanto, el primer nucleótido de adenina en el ARN GAPDH comienza a 300 pb (265 + 35). Limitamos arbitrariamente la longitud máxima de la cola de poly(A) a 250 (300 + 250 = 550). A partir de la tabla de regiones, el programa devuelve el tamaño promedio dentro de la región como 387 pb. - Utilice la ecuación (1) para calcular la longitud de la cola de poli(A) en el ARNm de interés:
Longitud de la cola de Poly(A) = (A - B - 35) (1)
Donde A es el promedio de pb del producto de PCR específico de poli(A) del electroferograma (es decir, 387 pb para GAPDH), B es el pb máximo del producto de PCR específico del gen del electroferograma (es decir, 265 pb para GAPDH) y "35" es la longitud de la etiqueta de cebador inverso universal.
NOTA: A partir del cálculo anterior, la longitud media de la cola de poli(A) de GAPDH es de 387 - 265 - 35= 87 pb.
10. Visualización de la distribución de la longitud de la cola de poli(A)
- Exportar datos como un archivo csv en Archivo | Exportación | Datos de ejemplo para recuperar datos de ejemplo.
NOTA: El archivo csv exportado muestra el tiempo de ejecución en lugar de bp en el eje X. - Vaya a un electroferograma de una muestra y marque Mostrar tamaños en la pestaña Electroferograma para convertir automáticamente la tabla de regiones en la tabla de regiones de bp a tiempo de ejecución. En el ejemplo, de 70,39 s a 86,28 s corresponden a 300 pb a 550 pb.
- Abra el archivo csv y seleccione los valores de 70,39 s a 86,28 s de tiempo de ejecución para generar un gráfico. Para visualizar los tamaños de pb en el eje X del gráfico, exporte el electroferograma con los tamaños de pb como un archivo de imagen y superpóngalo en el gráfico generado en la hoja de cálculo. Esto coincidirá adecuadamente con los tamaños de pb en la distribución de la cola de poli(A).
Access restricted. Please log in or start a trial to view this content.
Resultados
Aquí, analizamos la longitud de la cola de poli(A) de Dscam1 y GAPDH de cerebros de larvas de Drosophila (Figura 4). Los ARN aislados se visualizaron en un gel de agarosa para el control de calidad. Una sola banda de ARN con un tamaño de alrededor de 600 nucleótidos indica una preparación de ARN intacta (Figura 2A). Los ARN se sometieron a la electroforesis capilar de alta resolución y a la electroforesis capilar de alta resoluci?...
Access restricted. Please log in or start a trial to view this content.
Discusión
En este protocolo, describimos la técnica para diseccionar el cerebro de la larva de Drosophila de la etapa errante del3er estadio, así como la preparación de la muestra de células de Drosophila S2. Debido a la naturaleza lábil de los ARNm, la recolección de muestras requiere precaución adicional. Para la disección del cerebro de las larvas, los cerebros no deben dañarse durante el aislamiento y no deben mantenerse en solución durante un período prolongado. Es esencial mantener el ...
Access restricted. Please log in or start a trial to view this content.
Divulgaciones
Los autores no tienen conflictos de intereses que revelar.
Agradecimientos
Este estudio fue financiado por el Instituto Nacional de Trastornos Neurológicos y Accidentes Cerebrovasculares (National Institute of Neurological Disorders and Stroke Grant R01NS116463 to J.K., y el Centro de Imágenes Celulares y Moleculares de la Universidad de Nevada, Reno, que contó con el apoyo de los Institutos Nacionales de Salud Grant P20GM103650 y se utilizó para la investigación informada en este estudio.
Access restricted. Please log in or start a trial to view this content.
Materiales
| Name | Company | Catalog Number | Comments |
| 3-(N-morpholino) propanesulfonic acid (MOPS) | Research Product Internation (RPI) | M92020 | |
| Agilent High Sensitivity DNA Kit | Agilent Technologies | 5067-4626 | |
| Agilent software 2100 expert free download demo | Agilent Technologies | https://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-software/2100-expert-software-228259 | |
| Apex 100 bp-Low DNA Ladder | Genesee Scientific | 19-109 | |
| Bioanalyzer | Agilent 2100 Bioanalyzer G2938C | ||
| Diethyl pyrocarbonate (DEPC) | Research Product Internation (RPI) | D43060 | |
| DNA dye (Gel Loading Dye, Purple (6x) | New England biolabs | B7024S | |
| Drosophila S2 cell line | Drosophila Genomics Resource Center stock #181 | ||
| Drosophila Schneider’s Medium | Thermo Fisher Scientific | 21720024 | |
| Ehidium bromide | Genesee scientific | 20-276 | |
| Fetal bovine serum (FBS) | Sigma-Aldrich | F4135 | |
| Forceps Dumont 5 | Fine Science tools | 11254-20 | |
| Nuclease free water | Thermo Fisher Scientific | AM9932 | |
| PBS 10x | Research Product Internation (RPI) | P32200 | |
| Poly(A) Tail-Length Assay Kit | Thermo Fisher Scientific | 764551KT | |
| RiboRuler Low Range RNA Ladder | Thermo Fisher Scientific | SM1833 | |
| RNA Gel Loading Dye (2x) | Thermo Fisher Scientific | R0641 | |
| RNA microprep kit | Zymoresearch | R1050 | |
| RNA miniprep kit | Zymoresearch | R1055 | |
| Scissors-Vannas Spring Scissors - 2.5 mm Cutting Edge | Fine Science tools | 15000-08 | |
| TopVision Agarose Tablets | Thermo Fisher Scientific | R2802 | |
| Tris-Acetate-EDTA (TAE) | Thermo Fisher Scientific | B49 |
Referencias
- Stewart, M. Polyadenylation and nuclear export of mRNAs. Journal of Biological Chemistry. 294 (9), 2977-2987 (2019).
- Machida, K., et al. Dynamic interaction of poly(A)-binding protein with the ribosome. Scientific Reports. 8 (1), 17435(2018).
- Eisen, T. J., et al. The dynamics of cytoplasmic mRNA metabolism. Molecular Cell. 77 (4), 786-799 (2020).
- Liudkovska, V., Dziembowski, A. Functions and mechanisms of RNA tailing by metazoan terminal nucleotidyltransferases. Wiley Interdisciplinary Reviews RNA. 12 (2), e1622(2021).
- Goldstrohm, A. C., Wickens, M. Multifunctional deadenylase complexes diversify mRNA control. Nature Reviews Molecular Cell Biology. 9 (4), 337-344 (2008).
- Schmidt, M. J., Norbury, C. J. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. Wiley interdisciplinary reviews RNA. 1 (1), 142-151 (2010).
- Laishram, R. S. Poly(A) polymerase (PAP) diversity in gene expression - Star-PAP vs canonical PAP. FEBS Letters. 588 (14), 2185-2197 (2014).
- Salles, F. J., Lieberfarb, M. E., Wreden, C., Gergen, J. P., Strickland, S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 266 (5193), 1996-1999 (1994).
- Wreden, C., Verrotti, A. C., Schisa, J. A., Lieberfarb, M. E., Strickland, S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 124 (15), 3015-3023 (1997).
- Passmore, L. A., Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nature Reviews Molecular Cell Biology. 23 (2), 93-106 (2021).
- Murray, E. L., Schoenberg, D. R. Assays for determining poly(a) tail length and the polarity of mRNA decay in mammalian cells. Methods in Enzymology. 448, 483-504 (2008).
- Salles, F. J., Strickland, S. Analysis of poly(a) tail lengths by PCR: The PAT assay. Methods in Molecular Biology. 118, 441-448 (1999).
- Salles, F. J., Darrow, A. L., O'Connell, M. L., Strickland, S. Isolation of novel murine maternal mRNAs regulated by cytoplasmic polyadenylation. Genes and Development. 6 (7), 1202-1212 (1992).
- Salles, F. J., Strickland, S. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. Genome Research. 4 (6), 317-321 (1995).
- Janicke, A., Vancuylenberg, J., Boag, P. R., Traven, A., Beilharz, T. H. ePAT: A simple method to tag adenylated RNA to measure poly(a)-tail length and other 3' RACE applications. RNA. 18 (6), 1289-1295 (2012).
- Minasaki, R., Rudel, D., Eckmann, C. R. Increased sensitivity and accuracy of a single-stranded DNA splint-mediated ligation assay (sPAT) reveals poly(a) tail length dynamics of developmentally regulated mRNAs. RNA Biology. 11 (2), 111-123 (2014).
- Martin, G., Keller, W. Tailing and 3'-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA. 4 (2), 226-230 (1998).
- Kusov, Y. Y., Shatirishvili, G., Dzagurov, G., Verena, G. M. A new G-tailing method for the determination of the poly(a) tail length applied to hepatitis a virus RNA. Nucleic Acids Research. 29 (12), 57(2001).
- Bazzini, A. A., Lee, M. T., Giraldez, A. J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 336 (6078), 233-237 (2012).
- Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H., Bartel, D. P. Poly(a)-tail profiling reveals an embryonic switch in translational control. Nature. 508 (1), 66-71 (2014).
- Chang, H., Lim, J., Ha, M., Kim, V. N. TAIL-seq: Genome-wide determination of poly(a) tail length and 3' end modifications. Molecular Cell. 53 (6), 1044-1052 (2014).
- Legnini, I., Alles, J., Karaiskos, N., Ayoub, S., Rajewsky, N. FLAM-seq: Full-length mRNA sequencing reveals principles of poly(A) tail length control. Nature Methods. 16 (9), 879-886 (2019).
- Garalde, D. R., et al. Highly parallel direct RNA sequencing on an array of nanopores. Nature Methods. 15 (3), 201-206 (2018).
- Singh, M., Ye, B., Kim, J. H. Dual leucine zipper kinase regulates Dscam expression through a noncanonical function of the cytoplasmic poly(A)-binding protein. Journal of Neuroscience. 42 (31), 6007-6019 (2022).
- Macharia, R. W., Ombura, F. L., Aroko, E. O. Insects' RNA profiling reveals absence of "hidden break" in 28S ribosomal RNA molecule of onion thrips, Thrips tabaci. Journal of Nucleic Acids. 2015, 965294(2015).
- Miura, P., Sanfilippo, P., Shenker, S., Lai, E. C. Alternative polyadenylation in the nervous system: to what lengths will 3' UTR extensions take us. Bioessays. 36 (8), 766-777 (2014).
- Sement, F. M., et al. et al Uridylation prevents 3' trimming of oligoadenylated mRNAs. Nucleic Acids Research. 41 (14), 7115-7127 (2013).
Access restricted. Please log in or start a trial to view this content.
Reimpresiones y Permisos
Solicitar permiso para reutilizar el texto o las figuras de este JoVE artículos
Solicitar permisoExplorar más artículos
This article has been published
Video Coming Soon
ACERCA DE JoVE
Copyright © 2025 MyJoVE Corporation. Todos los derechos reservados