Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Investigating Tissue- and Organ-specific Phytochrome Responses using FACS-assisted Cell-type Specific Expression Profiling in Arabidopsis thaliana
W tym Artykule
Podsumowanie
The molecular basis of spatial-specific phytochrome responses is being investigated using transgenic plants that exhibit tissue- and organ-specific phytochrome deficiencies. The isolation of specific cells exhibiting induced phytochrome chromophore depletion by Fluorescence-Activated Cell Sorting followed by microarray analyses is being utilized to identify genes involved in spatial-specific phytochrome responses.
Streszczenie
Protokół
1. Plant Growth
- Confirmed UAS-BVR X GAL4-GFP enhancer trap line isolated as described4 (For summary see Fig. 1) and wild-type or parental lines are sown on soil, i.e. ~2000 sterilized seeds per line.
- Plants are grown for 5 weeks on soil under white illumination of 100 μmolm-2s-1 at 22 °C and 70% humidity.
2. Leaf Protoplast Isolation (adapted from Denecke and Vitale9)
- To isolate protoplasts, prepare TEX buffer. For 1 liter TEX buffer, weigh out the following components: 3.1 g Gamborg's B5 salts, 0.5 g 2-(N-morpholino)ethanesulfonic acid (MES, 2.56 mM), 0.75 g calcium chloride dihydrate (CaCl2.2H2O, 6.75 mM), 0.25 g ammonium nitrate (NH4NO3, 3.12 mM), 136.9 g sucrose (0.4M).Dissolve components completely in ~900 ml of deionized, distilled water (ddH2O) and bring the pH to 5.7 with 1 M KOH. Bring final volume to 1 liter and filter sterilize with a 0.2 μm bottle-top filter connected to a vacuum pump.
- Collect green, healthy leaves from plants (~250 ml of leaves loosely packed in a beaker) and rinse with~ 40 ml ddH2O 4 times, followed by rinsing twice with sterile ddH2O. Using a #20 scalpel, cut leaves into thin strips and divide tissue equally into two 50 ml sterile plastic tubes. Prepare 1x leaf digestion mixture by adding 5 ml aliquot of 10x leaf digestion stock solution (10x leaf digestion stock solution: dissolve 2% w/v Macerozyme R-10, 4% w/v Cellulase "Onozuka" R-10 in TEX buffer, filter sterilize with 0.2 μm filter and freeze 5 ml aliquots at -80 °C) to 45 ml fresh TEX buffer. Add ~25 ml of 1x leaf digestion mix per 50 ml tube to cover all leaf tissue.
- Vacuum infiltrate leaf tissue in digestion mixture in open tubes for 1 hr at room temperature using a vacuum desiccator connected to a water pump followed by 3-hr incubation at room temperature on a rocker with gentle shaking. Capped tubes with leaf tissue in 1x leaf digestion mix are kept wrapped in aluminum foil during this process to prevent exposure to light.
- After 3 hours, increase the rocker speed for ~2 minutes to release protoplasts. Filter the crude protoplast suspension through two layers of sterile cheese cloth to remove debris and collect the filtrate in a sterile glass beaker. Filter the filtrate through a sterile 100-μm nylon mesh into a sterile Petri dish. Collect the flow through and transfer it to a new sterile 50 ml tube. Use about 15-20 ml of fresh TEX buffer to wash the sterile Petri dish and collect any protoplasts adhering to the surface.
- Centrifuge the flow through using a swing bucket rotor at 100xg at 10 °C for 15 min (Acceleration 6, Deceleration 0).
- Remove ~25 - 30 ml of the liquid below the floating protoplast layer, which contains residual and pelleted debris, with a sterile 9" glass Pasteur pipette connected to a peristaltic pump without disturbing the floating protoplast layer and leaving ~10 - 15 ml volume.
- Add fresh TEX buffer to a final volume of 40 ml while gently resuspending the protoplasts.
- Repeat steps 2.5 to 2.7 two times to remove as much cellular debris as possible. The centrifugation time is reduced to 10 minutes in the first repetition and to 5 minutes in the final repetition.
- Aspirate floating protoplasts with a cut, sterile 1 ml transfer pipette into a new 15 ml tube. Proceed directly to sorting or store in dark for up to 2 hours at 4 °C until sorting.
3. Protoplast Sorting by Fluorescence-Activated Cell Sorting (FACS)
- Before sorting, examine protoplasts by Confocal Laser Scanning Microscopy (CLSM) using a 488-nm laser for excitation to confirm protoplast integrity and the presence of GFP fluorescence in the protoplast pool. Minimal debris is necessary to avoid clogging the FACS sorting nozzle.
- Sort isolated protoplasts in TEX buffer via FACS (BD FACSVantage SE, BD Biosciences) using a 200-μm nozzle on a macro sort head at event rates between 6,000 and 15,000, with a system pressure of around 9 p.s.i. following an adapted protocol10.
- Wild-type non-GFP protoplasts are used to determine the autofluorescence thresholds.
- To collect GFP-positive protoplasts, sort cells using an air-cooled argon laser (Spectra Physics Model 177, Newport Corporation, Irvine, CA) operated at 100 mW on a 488-nm argon line to identify GFP fluorescence using a 530/30 band pass filter.
- Following the collection of GFP-positive protoplasts, examine sorted protoplasts by CLSM as detailed in step 3.1.
- Extract total RNA from sorted protoplasts using a Qiagen RNeasy Plant Mini Kit. cDNA can then be prepared for hybridization as described in Affymetrix GeneChip Expression Analysis Technical Manual and then hybridized to AtH1 Arabidopsis Affymetrix whole genome arrays.
Representative Results
We detected a significant number of GFP-positive cells in the GFP channel by FACS (Fig. 2). In optimization assays using GFP enhancer-trap parents, the constitutively GFP-expressing line, J0571, displayed ~ 17% to 24% GFP-positive protoplasts, whereas the line with vascular and dermal expression, J1071, had ~1.4% GFP-positive protoplasts (Table 1). Sorting of 3 x 500 μl (~ 1.5 ml total of protoplast suspension) of J0571 protoplasts for 1 h gave ~ 100,000 GFP-positive protoplasts. Sorting of 3 x 500 μl (~ 1.5 ml total protoplast suspension) of J1071 protoplasts for 1.5 h gave ~ 3,000 GFP-positive protoplasts. Confocal images indicated a very high yield of protoplasts for both J0571 and J1071 samples before sorting was carried out (Fig. 3C and 3E), and the sorted fractions contained only bright GFP-fluorescent protoplasts (Fig. 3G and data not shown). This finding confirms that intact GFP-positive protoplasts can be sorted via FACS. The non-GFP protoplasts can also be sorted and collected in a separate channel. The non-GFP protoplasts serve as an ideal negative control for subsequent microarray analyses to detect the specific changes in gene expression upon reduction of holophytochrome levels. RNA extraction from isolated protoplasts (1 ml) yields RNA of sufficient quantity for detection by fluorospectrometry (Fig. 4). Isolated RNA was assessed using a NanoDrop instrument (NanoDrop 1000, Thermo Scientific) and quantified by NanoDrop 3.7.1 RNA quantification software. RNA yields from pre-sorted C24 wild-type protoplasts or FACS-sorted, GFP-positive enhancer trap protoplasts exceeded the minimum 20 ng needed for use in RNA-labeling assays for microarray (Table 2).
| Enhancer Trap Line | % of GFP-positive protoplasts | Number of sorted GFP protoplasts |
| J0571 | 17.18 % ~ 24.06 % | 26,400 ~ 36,000 |
| J1071 | ~ 1.43 % | 1,000 ~ 1,300 |
Table 1. Percentage of GFP-positive protoplasts from two enhancer trap lines before sorting and the number of GFP-positive protoplasts collected by running 500 μL of protoplast suspension through the Fluorescence Activated Cell Sorter (FACSVantage, BD).
| Plant Line | RNA yield (ng) |
| C24 WT | 12486.5 |
| J0571 | 60 |
| J1071 | 71.5 |
Table 2. Yield from RNA isolation from protoplasts. RNA was isolated from pre-sorted C24 wild-type or sorted GFP-positive protoplasts from two enhancer trap lines and quantified.
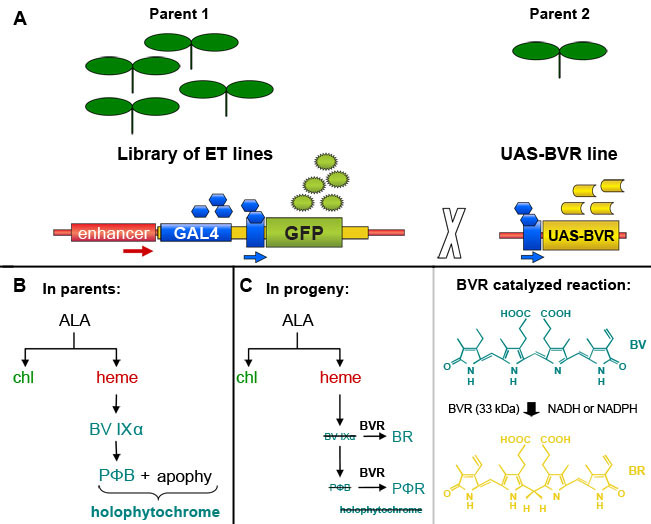
Figure 1. GAL4 enhancer-trap-based induction of Biliverdin Reductase (BVR) expression in transgenic Arabidopsis thaliana plants. (A). An individual selected from a library of GAL4-based enhancer trap lines, which contain a GAL4-responsive GFP marker gene, can be crossed with a line containing a GAL4-responsive target gene to induce expression of the target gene in GAL4-containing cells marked by GFP fluorescence. Based on a figure from Dr. Jim Haseloff (http://www.plantsci.cam.ac.uk/Haseloff/geneControl/GAL4Frame.html). (B) Production of phytochrome chromophore, phytochromobilin (PΦB) and holophytochrome in parent lines. (C) Left, Reduction of biliverdin IXα (BV IXα) and PΦB by biliverdin reductase (BVR) activity to BR and PΦR, respectively. BVR activity results in depletion of PΦB and leads to a reduction in the production of photoactive holophytochrome. Right, the reaction catalyzed by BVR is shown.
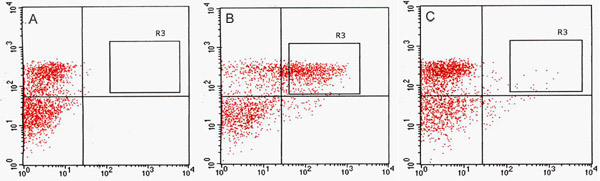
Figure 2. Fluorescence Activated Cell Sorting (FACS) acquisition dot plots. Comparison of protoplast cell sorting for C24 wild type (A), J0571 (B) and J1071 (C). (A) Acquisition dot plot of non-GFP fluorescent C24 wild-type protoplasts excited by a 488-nm argon laser and used to determine autofluorescence threshold. (B) and (C) acquisition dot plots show proportions of protoplasts that are GFP positive in response to excitation by a 488-nm laser. R3 sorting gates in B and C delimit the GFP-positive targets that were sorted by Fluorescence-Activated Cell Sorter (FACSVantage, BD) and collected. Red channel indicates values for chlorophyll autofluorescence from protoplasts and green channel indicates values for GFP fluorescence.

Figure 3. Confocal microscopy of plant protoplasts used for cell sorting. Confocal laser scanned images of protoplasts before (A, C, E) and after (G) sorting via Fluorescence-Activated Cell Sorter (FACS). B, D, F and H are DIC images. Protoplasts of C24 wild type (A, B), J1071 (C, D) and J0571 (E, F, G, H) are shown. Images C, E and G are merged images of GFP fluorescence (BP 505 nm - 575 nm) and Autofluorescence (LP 650 nm) obtained from excitation with a 488-nm laser. A through H are average of 4 scans under 63x oil. Bar = 10 μm.
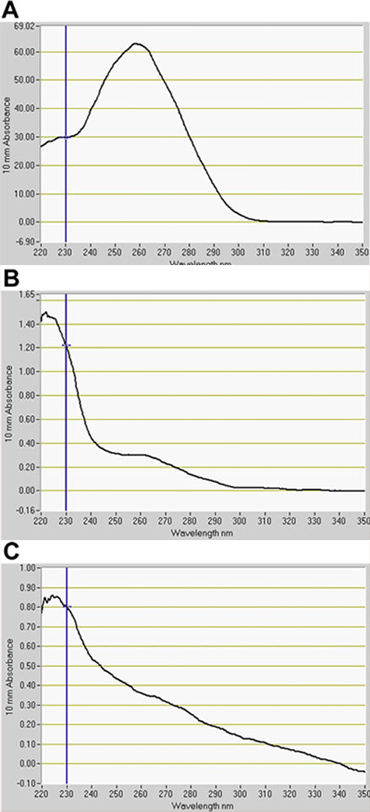
Figure 4. Quantification of RNA isolated from protoplasts by fluorospectrometry. RNA extracted from (A) C24 wild type, (B) J0571, and (C) J1071. A indicates RNA for pre-sorted wild-type protoplasts. B and C indicate RNA from GFP-positive protoplasts sorted by Fluorescence-Activated Cell Sorting (FACS). RNA quantification software, NanoDrop 3.7.1, (NanoDrop 1000, Thermo Scientific).
Dyskusje
Gene expression profiling through microarrays (1) has indicated that more than 30% of the genes in Arabidopsis seedlings are light regulated11 and (2) has identified a vast group of genes encoding light signal transduction proteins involved in the phytochrome signaling cascade12, 13. Such experiments suggest that light induces rapid and long-term changes in gene expression. Each pool of phytochromes may control only a subset of developmental and adaptive responses. Moreover, it is likely that downst...
Podziękowania
Work in the Montgomery lab on phytochrome responses in plants is supported by the National Science Foundation (grant no. MCB-0919100 to B.L.M.) and the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (grant no. DE FG02 91ER20021 to B.L.M.). We thank Melissa Whitaker for technical assistance during filming and critically reading the manuscript, Stephanie Costigan for experimental assistance, Dr. Louis King for assistance with developing and optimizing Fluorescence-Activated Cell Sorting protocols for Arabidopsis protoplast sorting and Dr. Melinda Frame for assistance with confocal microscopy. We thank Marlene Cameron for graphical design assistance and Karen Bird for editorial assistance.
Materiały
| Name | Company | Catalog Number | Comments |
| Anti-BVR antibody | QED Bioscience Inc. | 56257-100 | |
| Cellulase “Onozuka” R-10 | SERVA Electrophoresis | MSPC 0930 | |
| Gamborg’s B5 basal salt mixture | Sigma-Aldrich | G5768 | |
| Macerozyme R-10 | SERVA Electrophoresis | PTC 001 | |
| MES, low moisture content | Sigma-Aldrich | M3671 | |
| Murashige and Skoog salts | Caisson Laboratories | 74904 | |
| Phytablend | Caisson Laboratories | 28302 | |
| RNeasy Plant Minikit | Qiagen | 16419 |
Odniesienia
- Franklin, K. A., Quail, P. H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11-24 (2010).
- Montgomery, B. L. Right place, right time: Spatiotemporal light regulation of plant growth and development. Plant Signal Behav. 3, 1053-1060 (2008).
- Laplaze, L. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 56, 2433-2442 (2005).
- Costigan, S., Warnasooriya, S. N., Montgomery, B. L. Root-localized phytochrome chromophore synthesis is required for tissue-specific photoregulation of root elongation and impacts sensitivity to jasmonic acid in Arabidopsis thaliana. , .
- Lagarias, D. M., Crepeau, M. W., Maines, M. D., Lagarias, J. C. Regulation of photomorphogenesis by expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Cell. , 675-688 (1997).
- Montgomery, B. L., Yeh, K. C., Crepeau, M. W., Lagarias, J. C. Modification of distinct aspects of photomorphogenesis via targeted expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Physiol. 121, 629-639 (1999).
- Warnasooriya, S. N., Montgomery, B. L. Detection of spatial-specific phytochrome responses using targeted expression of biliverdin reductase in Arabidopsis. Plant Physiol. 149, 424-433 (2009).
- Warnasooriya, S. N., Porter, K. J., Montgomery, B. L. Light-dependent anthocyanin accumulation and phytochromes in Arabidopsis thaliana. , .
- Denecke, J., Vitale, A. The use of protoplasts to study protein synthesis and transport by the plant endomembrane system. Methods Cell Biol. 50, 335-348 (1995).
- Birnbaum, K. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods. 2, 615-619 (2005).
- Ma, L. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 13, 2589-2607 (2001).
- Chen, M., Chory, J., Fankhauser, C. Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87-117 (2004).
- Ulm, R., &, N. a. g. y., F, . Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant Biol. 8, 477-482 (2005).
- Ma, L. Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138, 80-91 (2005).
- Neff, M. M., Fankhauser, C., &, C. h. o. r. y., J, . Light: an indicator of time and place. Genes Dev. 14, 257-271 (2000).
- Birnbaum, K. A gene expression map of the Arabidopsis root. Science. 302, 1956-1960 (2003).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone