Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
In Vitro Evaluation of Oncogenic Transformation in Human Mammary Epithelial Cells
W tym Artykule
Podsumowanie
This protocol provides experimental in vitro tools to evaluate the transformation of human mammary cells. Detailed steps to follow-up cell proliferation rate, anchorage-independent growth capacity, and distribution of cell lineages in 3D cultures with basement membrane matrix are described.
Streszczenie
Tumorigenesis is a multi-step process in which cells acquire capabilities that allow their growth, survival, and dissemination under hostile conditions. Different tests seek to identify and quantify these hallmarks of cancerous cells; however, they often focus on a single aspect of cellular transformation and, in fact, multiple tests are required for their proper characterization. The purpose of this work is to provide researchers with a set of tools to assess cellular transformation in vitro from a broad perspective, thereby making it possible to draw sound conclusions.
A sustained proliferative signaling activation is the major feature of tumoral tissues and can be easily monitored under in vitro conditions by calculating the number of population doublings achieved over time. Besides, the growth of cells in 3D cultures allows their interaction with surrounding cells, resembling what occurs in vivo. This enables the evaluation of cellular aggregation and, together with immunofluorescent labeling of distinctive cellular markers, to obtain information on another relevant feature of tumoral transformation: the loss of proper organization. Another remarkable characteristic of transformed cells is their capacity to grow without attachment to other cells and to the extracellular matrix, which can be evaluated with the anchorage assay.
Detailed experimental procedures to evaluate cell growth rate, to perform immunofluorescent labeling of cell lineage markers in 3D cultures, and to test anchorage-independent cell growth in soft agar are provided. These methodologies are optimized for Breast Primary Epithelial Cells (BPEC) due to its relevance in breast cancer; however, procedures can be applied to other cell types after some adjustments.
Wprowadzenie
Multiple successive events are required for neoplasm development. In 2011, Hanahan and Weinberg described 10 capabilities that enable transformed cells’ growth, survival, and dissemination: the so-called “Hallmarks of Cancer”1. The methodology described here compiles three different tools to evaluate in vitro cellular transformation by focusing on some of the tumoral cells’ distinctive features. These techniques assess the cell proliferation rate, the behavior of cells when cultured in 3D and their capacity to form colonies with anchorage independence.
Cell models are crucial to test hypothesis in vitro. Different approaches have been developed to generate experimental models of cellular transformation for the study of cancer2,3,4. Since breast cancer is the most common cancer among women worldwide and is responsible for approximately 15% of cancer deaths among women5, providing suitable cellular models of mammary epithelial cells is of utmost importance for further investigation. In this article, we have illustrated the potential of three techniques to evaluate cellular transformation using an experimental model of Breast Primary Epithelial Cells (BPECs) transformation initially described by Ince and colleagues in 20076 and later implemented in our laboratory7. This experimental model is based on the sequential alteration of three targeted genes (SV40 Large T and small t antigens herein referred to as Ttag, hTERT, and HRAS) to the genome of non-transformed BPECs. Moreover, the method used for BPECs derivation favors the maintenance of mammary epithelial cells with luminal or myoepithelial markers, resulting in a heterogeneous cell culture that retains some of the mammary gland physiological traits.
In the mammary gland, luminal mammary epithelial cells, which are responsible for milk production, are located near the lumen, whereas myoepithelial cells are disposed around luminal cells and take care of contraction movements leading the milk to the nipple. The loss of proper organization between these cell lineages is a feature of tumoral transformation8 that can be assessed in vitro after immunofluorescent detection of distinctive lineage markers in 3D cell cultures. Another major characteristic of tumoral cells is their capacity to grow without attachment to other cells and to the extracellular matrix1. When healthy cells are forced to grow in suspension, mechanisms such as anoikis ‒ a type of cell death induced in response to detachment from the extracellular matrix ‒ are activated9. The evasion of cell death is one of the distinctive hallmarks of cancer and thus, transformed cells are capable to inactivate anoikis and survive in an anchor-independent manner. This capacity can be evaluated in vitro with the anchorage-independent assay using soft agar. Furthermore, an inherent feature of tumoral tissues is their sustained proliferative signaling capacity, which can be easily monitored under in vitro conditions by measuring the increase of cell number along time, not only in suspension assays but also by monitoring the growth rate of monolayer adherent cultures.
Despite the best model to test tumorigenic potential is the inoculation of tumoral cells in murine models and evaluation of tumor development in situ, it is important to minimize the number of animals employed in experimental procedures as much as possible. Therefore, having suitable tests to assess transformation in vitro is a top priority. Here, we provide a set of tools to evaluate the tumorigenic potential of partially and fully transformed breast epithelial cells that can be easily implemented in most of the laboratories that work with cellular transformation models.
Protokół
Human samples used in the following experiments were obtained from reduction mammoplasties carried out at Clínica Pilar Sant Jordi (Barcelona) under standard procedure consent. All procedures are performed in a Class II Biological Safety Cabinet unless otherwise stated.
1. In vitro culture of human mammary epithelial cells and growth curve plot build-up
- In vitro culture of breast primary epithelial cells (BPECs): cell passaging
NOTE: For BPEC derivation and cell culture follow instructions described by Ince et al., 20076.- Medium preparation.
- Supplement WIT basal defined medium with P or T supplements, provided by the manufacturer, depending on whether primary or transformed BPECs are cultured.
- Add cholera toxin to the supplemented WIT medium to a final concentration of 100 ng/mL for primary or 25 ng/mL for transformed BPECs.
CAUTION: Cholera toxin is fatal if swallowed. Use personal protective equipment. Avoid its release to the environment.
- Cell culture maintenance and passaging.
NOTE: For the following steps keep in mind that cells are growing in a T25 flask. Nonetheless, volumes can be adapted to other cell culture formats maintaining the proportionality in terms of surface area.- Check on the cell confluency every day. When the culture is 90% confluent, perform cell passaging.
- Acquire 1x PBS, 3x trypsin, medium and a 15 mL conical tube containing 2 mL of Fetal Bovine Serum (FBS) for each flask.
- Remove the medium from the flask and keep it in the 15 mL conical tube containing FBS.
- Rinse cells with 1x PBS.
- Detach cells from the surface by adding 1 mL of 3x trypsin. Incubate for 5 min at 37 ˚C.
- Check if cells have been detached. Apply vigorous shaking if cells are not completely detached.
- Inactivate trypsin by adding the reserved medium supplemented with FBS.
- Harvest the cellular suspension and place it in the 15 mL conical tube.
- Centrifuge at 500 x g for 5 min, eliminate the supernatant, and resuspend the pelleted cells by flicking the bottom of the tube with a finger.
- Add 1–2 mL of fresh media to the pellet and measure the cell concentration using an automatic cell counter or a hemocytometer. This data will later be used to calculate the population doublings and draw the growth curve. Seed 12,000 cells/cm2 (e.g., 300,000 cells for a T25 flask) in modified cell culture surface flasks (see Table of Materials).
NOTE: Dilute the cell suspension solution if the concentration is too high to ensure proper quantification. - Add medium to a final volume of 5 mL and incubate the cells at 37 ˚C and 5% CO2 atmosphere.
- Replace the cell culture medium every 48 h.
- Medium preparation.
- Population doubling calculation and data visualization
- Using data of the cell count obtained in the step 1.1.2.10, apply the following formula to obtain the accumulated population doubling (PD) values:
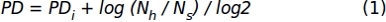
Where, PDi denotes the number of population doublings achieved by the cells until the previous subculture (it refers to the PD accumulated on the previous subculture), Nh is the number of harvested cells, and Ns is the number of seeded cells. - Represent data for a specific interval of time using an XY graph where the number of days in culture (x-axis) and the accumulated PD (y-axis) are represented.
- Get the best-fit line and the fitting equation:

NOTE: An increased slope (b) means an increased proliferation rate.
- Using data of the cell count obtained in the step 1.1.2.10, apply the following formula to obtain the accumulated population doubling (PD) values:
2. Three-dimensional (3D) culture in basement membrane matrix and immunofluorescent protein detection
- 3D culture in basement membrane matrix
NOTE: This protocol has been adapted from Debnath et al., 200310 and is optimized for 24 well plates (see Table of Materials).- Prepare the material a day before the experiment: pre-chill basement membrane matrix overnight at 4 ˚C and let pipette tips, microcentrifuge tubes, and well plates cool in the freezer.
NOTE: The matrix must be kept at -20 ˚C for long-term storage. Make aliquots to avoid multiple freeze-thaw cycles. - On the day of the experiment, place pre-cooled material on ice.
- Rinse wells with cold sterile 1x PBS in order to reduce surface tension.
- Cover the bottom of each well with 100 µL of basement membrane matrix.
NOTE: Dispense the matrix slowly and spread it throughout the well; it is crucial to avoid bubble formation in the bottom layer to avert monolayer cell culture growth. - Place the plate in the incubator, at 37 ˚C, to let the matrix layer solidify.
NOTE: It usually takes about 20 min to solidify. - Meanwhile, trypsinize cells as explained previously in step 1.1. Centrifuge the cells at 500 x g for 5 min and resuspend in medium. Prepare a 400,000 cells/mL suspension and gently disaggregate any cell clump by pipetting.
- Prepare the medium with 8% basement membrane matrix and mix 1:1 (v/v) with cellular suspension to obtain a 200,000 cells/mL solution in 4% matrix.
NOTE: Calculate the amount of medium needed to avoid matrix unnecessary waste. - Place 500 µL of cell suspension in matrix solution on top of the already solidified matrix layer to seed a total amount of 100,000 cells in medium with 4% basement membrane matrix.
- Incubate cells at 37 ˚C for a few minutes and then, add 500 µL of the medium with 4% basement membrane matrix. Incubate the cells at 37 ˚C in an incubator with 5% CO2 for 14 days. Seeded cells will group and proliferate to originate the acini-like structures.
NOTE: Cell motility and aggregation can be monitored by time-lapse during the 3D formation process. Use image analysis software (e.g., Fiji/ImageJ or Imaris) to evaluate these events. The number and size of the acini depend on the aggregation process and the proliferation rate and might vary between cell types. Adjust basement membrane matrix concentration and seeded cells to obtain desired 3D structures. - Add 500 µL of the medium with 4% basement membrane matrix 2–3 times per week.
NOTE: Avoid disturbance of the layers carrying the plate gently during manipulation. - If desired, the number and size of acini can be measured during the culture period. To do so, take random pictures at different times after seeding using a phase contrast or DIC inverted microscope. Use image analysis software to measure the diameter of 100–200 3D structures.
- Prepare the material a day before the experiment: pre-chill basement membrane matrix overnight at 4 ˚C and let pipette tips, microcentrifuge tubes, and well plates cool in the freezer.
- Immunostaining
NOTE: Sterile conditions are not required during this part of the protocol.- Remove the culture medium.
- Tear the basement membrane matrix using a p200 pipette tip with the end cut off. Place ~50 µL of disaggregated matrix on top of a glass slide and smear it in an area of 1–2 cm2.
- Let the sample dry completely at room temperature or use a heating plate at 37 ˚C to accelerate the process. Fix samples with methanol:acetone (1:1, v/v) at -20 ˚C for 30 min.
NOTE: Fluorescent signal of previous markers, such as that of fluorescent proteins expressed by the cells, will be erased.
CAUTION: Methanol is flammable, toxic if inhaled, swallowed or in case it comes in contact with skin. Wear personal protective equipment and work inside a fume hood. - Discard the fixation solution and remove the excess, if any, by reclining the slide on filter paper.
NOTE: The protocol can be paused here. Once dry, slides can be stored at -20 ˚C for several months. - Block samples epitopes with 5% normal goat serum and 0.1% triton-X-100 in 1x PBS (blocking solution) for 2 h at room temperature.
- Meanwhile, prepare antibodies working solutions by diluting primary or secondary antibodies at the desired concentration in blocking solution.
NOTE: Antibody concentration must be accurately adjusted depending on the cell type and the antibody reference. As a guide, to identify cells from luminal and myoepithelial lineages in BPEC, primary anti-Cytokeratin 14 and anti-Claudin-IV antibodies (see Table of Materials) can be used. The recommended working solution concentration is 1:100 for these primary antibodies and 1:500 for anti-Mouse and anti-Rabbit secondary antibodies (see Table of Materials). - Add 30 µL of primary antibodies working solution and cover it with a strip of laboratory wrapping film to avoid evaporation. Incubate overnight at 4 ˚C in a humid chamber.
- Wash three times with 1x PBS for 1 h each.
- Repeat step 2.2.7 for secondary antibodies. Incubation should be performed in darkness.
- Wash with 1x PBS for 2 h.
NOTE: Adjust antibodies concentration, incubation time, and washing hardness to improve the signal/noise ratio for specific samples. - Remove the remaining PBS and, once dry, counterstain with DAPI at 0.25 µg/mL diluted in antifade mounting medium. Cover slides with a coverslip by letting it settle without applying pressure. Seal with nail polish.
NOTE: Samples can be stored at 4 ˚C for several weeks. For long-term storage, keep them at -20 ˚C. - Analyze fluorescent signal distribution for each acinus using a confocal microscope.
NOTE: Confocal microscope configuration must be accurately determined depending on the equipment used and the antibodies applied to the sample. As a guide, with the equipment and reagents detailed in the Table of Materials, use a 40x objective and the following laser and detector settings: for DAPI use excitation with a 405 laser (3%–5%), detection with a PMT detector (800V, Offset: -9) and a spectral band from 410 nm to 500 nm; for A488 (Claudin-IV) use excitation with a 488 laser (7%–10%), detection with a PMT detector (800V, Offset: -20) and a spectral band from 490 nm to 550 nm; and for Cy3 (Cytokeratin 14) use excitation with a 555 laser (2%–10%), detection with a PMT detector (800V, Offset: -35) and a spectral band from 560 nm to 600 nm.
3. Anchorage-independent assay, MTT staining and automatic colony quantification
- Anchorage-independent assay: agar and cellular suspension plating
NOTE: Protocol has been adapted from Borowicz et al., 201411 to perform experiments in BPECs.- Prepare a 1.2% agar solution diluted in ultrapure water in a sterile bottle. Autoclave the solution and maintain it at 42 ˚C during the experiment. The agar solution can be stored at 4 ˚C; when required, heat the agar solution until it is liquid again.
CAUTION: Use heat-resistant gloves to avoid burn after autoclave.
NOTE: From now on, sterile conditions must be maintained. - Prepare a 0.6% agar solution by mixing 1:1 (v/v) complete pre-warmed medium with 1.2% agar solution. Maintain at 42 ˚C to avoid premature solidification.
NOTE: Medium can be previously double supplemented to obtain a fully supplemented 0.6% agar + medium solution once mixed. - Cover the bottom of a 35 mm well with 1.5 mL of 0.6% agar in medium solution and let it solidify at room temperature. Make sure that the bottom of the plate is completely covered before agar solidification, otherwise, cells may adhere to the plate and grow in monolayer.
NOTE: Adherent and non-adherent surface plates can be used. - Meanwhile, trypsinize cells and, once centrifuged and resuspended in medium, prepare a 50,000 cells/ml solution and gently disaggregate any cell clump by pipetting repeatedly.
- Prepare a 0.3% agar + cell suspension in the medium at a final concentration of 25,000 cells/mL.
NOTE: Optimal cell concentration may differ among cell types. Try different concentrations until individualized colonies are formed.- Place a 40 µm strainer filter on top of a 50 mL sterile tube and filter the 50,000 cells/mL solution letting it drop into the bottom of the tube.
- Remove the filter from the 50 mL sterile tube, tilt the cell-containing tube to a 45˚ angle and drop the same volume of 0.6% agar + medium solution pouring it through the internal wall of the tube. This will allow the agar solution to cool down just enough to not damage the cells and avoid its premature solidification.
- Homogenize the mixture and deposit 1 mL of 0.3% agar + cell suspension in the medium (containing 25,000 cells) on top of the previously solidified bottom agar layer.
- Visualize seeded cells using an inverted microscope to make sure that the cells are individualized. Otherwise, the experiment should be repeated.
- Wait until the agar layer is completely solidified, then carefully add 1 mL of the fresh medium on top without disturbing the delicate agar layers beneath.
- Incubate the cells at 37 ˚C and 5% CO2 in an incubator for 3 weeks.
NOTE: The time required for colony formation can vary among different cell types, but usually 3 weeks are sufficient. - Change medium twice per week. To do so, gently tilt the plate toward you, aspirate medium in the lower corner, and add 1 mL of fresh medium.
NOTE: Avoid touching the agar layers as they easily detach from the plate.
- Prepare a 1.2% agar solution diluted in ultrapure water in a sterile bottle. Autoclave the solution and maintain it at 42 ˚C during the experiment. The agar solution can be stored at 4 ˚C; when required, heat the agar solution until it is liquid again.
- MTT staining
- Prepare Thiazolyl Blue Tetrazolium Bromide (MTT) stock solution at 6 mg/mL in ultrapure water in a sterile bottle and filter solution using 0.2 µm filters. This MTT solution can be stored for up to 6 months at -20 ˚C.
CAUTION: MTT may cause irritation and is suspected of causing genetic defects. Use safety glasses, gloves, and a respiratory filter.
NOTE: Avoid repeated freeze-thaw cycles. - Prepare a working solution of MTT at 1 mg/mL by diluting the stock solution with sterile ultrapure water.
- Once the colony formation period has concluded, remove the medium from the plate and add 1 mL of 1 mg/mL MTT to each well.
- Incubate for 24 h in the incubator. Remove the MTT solution by aspirating it gently. Plates can be stored at 4 ˚C for several weeks.
NOTE: Avoid light exposure to prevent non-specific crystal formation.
- Prepare Thiazolyl Blue Tetrazolium Bromide (MTT) stock solution at 6 mg/mL in ultrapure water in a sterile bottle and filter solution using 0.2 µm filters. This MTT solution can be stored for up to 6 months at -20 ˚C.
- Colony quantification
- Obtain images of each plate using an inverted microscope. Adjust the magnification in order to acquire the maximum field of view with fewer images and still being able to detect small colonies (usually 4x or 10x objectives).
NOTE: Make sure that images present a homogeneous background. Neither phase-contrast nor differential interference contrast is required since non-stained colonies will not be quantified. - Upload images to ImageJ/Fiji software12 to count the number of colonies and the area of each MTT-positive colony.
NOTE: A script for automatic quantification is provided as a Supplementary File. To execute the code, paste it to the macro editor (Plugins | New | Macro) and follow the instructions.- Obtain a binary mask through thresholding the original image (Image | Adjust | Threshold) to obtain well-delimited colonies (Figure 1).
NOTE: 8- or 16-bit images are usually required to perform this step. The “Minimum threshold” method is recommended. - Run the Extended Particle Analyzer from Biovoxxel plugin13 (Plugins | BioVoxxel | Extended Particle Analyzer) to identify MTT positive colonies (Figure 2). Initial guiding conditions: Size (µm2) = 250–Infinity; Solidity = 0.75–1.00.
- Obtain a binary mask through thresholding the original image (Image | Adjust | Threshold) to obtain well-delimited colonies (Figure 1).
- Estimate the average diameter (D) from each colony area value (A) according to the formula:

- Filter results by excluding low proliferative colonies (e.g., low diameter).
- Choose a minimum number of divisions per week (m) to be considered (e.g., 1).
- Estimate the radius of a colony (R) with n cells according to the following formula:

Where, r is the average radius of individual cells in suspension, n is the number of cells forming a colony that suffered m divisions each week during w weeks in culture. In exponential growth: n = 2(m*w). ρ is the Packaging efficiency. Note that, in random movement, packaging efficiency is ~0.64 and the densest possible packing fraction for identical spheres is 0.7414. - Discard all colonies that present a diameter lower than 2R as their cells have not achieved the minimum number of divisions considered in step 3.3.4.1.
- Obtain images of each plate using an inverted microscope. Adjust the magnification in order to acquire the maximum field of view with fewer images and still being able to detect small colonies (usually 4x or 10x objectives).
Wyniki
An experimental model of cellular transformation with the introduction of three genetic elements in BPECs was chosen to generate representative results of oncogenic transformation6,7 (Figure 3). Non-transformed BPECs (N) were derived from disease-free breast tissue as described by Ince and colleagues6 and cultured following the protocol indicated here. After overcoming STASIS (stress ...
Dyskusje
The experimental protocols described in this paper provide useful tools to assess the oncogenic transformation of in vitro cultured cells. Each technique evaluates specific aspects of the transformation process, and thus, special attention must be paid when drawing conclusions from a single analysis. Growth curves build-up is an approach that demands information already available when culturing cells for other purposes. That makes this technique cheaper and easier to apply compared to other cell proliferation assays. How...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
The AG laboratory is funded by the Spanish Nuclear Safety Council. T.A. and A.G. are members of a research group recognized by Generalitat de Catalunya (2017-SGR-503). MT holds a contract funded by the Scientific Foundation Asociación Española Contra el Cáncer [AECC-INVES19022TERR]. G.F. contract is funded by a grant from Cellex Foundation.
Materiały
| Name | Company | Catalog Number | Comments |
| 1 ml Serological Pipettes | Labclinics | PLC91001 | |
| 1.5 ml Eppendorfs | Thermo Fisher Scientific | 3451 | Dark eppendorfs are preferred for MTT long-term storage |
| 10 μl Pipette tips w/o filter | Biologix | 20-0010 | |
| 100 ml glass bottle | With cap, autoclavable | ||
| 1000 μl Pipette tips w/ filter | Labclinics | LAB1000ULFNL | |
| 1000 μl Pipette tips w/o filter | Biologix | 20-1000 | |
| 15 ml Conical tubes | VWR | 525-0400 | |
| 2 ml Serological Pipettes | Labclinics | PLC91002 | |
| 200 μl Pipette tips w/ filter | Labclinics | FTR200-96 | |
| 5 ml Serological Pipettes | Labclinics | PLC91005 | |
| 50 ml Conical Tubes | VWR | 525-0304 | |
| Acetone | PanReac AppliChem | 211007 | Used for 3D structure fixation prior to immunofluorescent labelling |
| Agar | Sigma-Aldrich | A1296 | Used for anchorage assay |
| Anti-Claudin 4 antibody | Abcam | 15104, RRID:AB_301650 | Working dilution 1:100, host: rabbit |
| Anti-Cytokeratin 14 [RCK107] antibody | Abcam | 9220, RRID:AB_307087 | Working dilution 1:100, host: mouse |
| Anti-mouse Cyanine Cy3 antibody | Jackson ImmunoResearch Inc. | 115-165-146, RRID:AB_2338690 | Working dilution 1:500, host: goat |
| Anti-rabbit Alexa Fluor 488 antibody | Thermo Fisher Scientific | A-11034, RRID:AB_2576217 | Working dilution 1:500, host: goat |
| Autoclave | |||
| BioVoxxel Toolbox | RRID:SCR_015825 | ||
| Cell culture 24-well Plate | Labclinics | PLC30024 | Used for 3D cultures in Matrigel. Flat Bottom |
| Cell culture 6-well Plate | Labclinics | PLC30006 | Used for anchorage assay |
| Cell incubator (37 ºC and 5 % CO2) | |||
| Cell Strainers | Fisherbrand | 11587522 | Mesh size: 40 μm |
| CellSense software | Olympus | Used to image acquisition | |
| Centrifuge | |||
| Cholera Toxin from Vibrio cholerae | Sigma-Aldrich | C8052 | Used to supplement cell culture medium |
| Class II Biological Safety Cabinet | Herasafe | HAEREUS HS12 | |
| Confocal inverted Microscope | Leica | TCS SP5 | |
| Cover glasses | Witeg Labortechnik GmbH | 4600122 | 22 X 22 mm, thickness 0.13 - 0.17 mm |
| DAPI | 2-(4-amidinophenyl)-1H -indole-6-carboxamidine | ||
| Fetal Bovine Serum | Biowest | S1810 | Used to inactivate trypsine action |
| Fiji software (ImageJ) | National Institutes of Health | RRID:SCR_002285 | Free download, no license needed |
| Glass Pasteur Pipettes | |||
| Glass slides | Fisherbrand | 11844782 | |
| Goat Serum | Biowest | S2000 | Used for immunofluorescence of 3D structures |
| Heat-Resistant Gloves | Used for agar manipulation after autoclave | ||
| Heater bath (37 ºC) | Used to temper solutions prior to cell subculture | ||
| Heater bath (42 ºC) | Used to keep agar warm | ||
| Heating plate | Used for Matrigel dehydration | ||
| Humid chamber | Used for the incubation of antibodies during immunofluorescence | ||
| Ice | Used during Matrigel manipulation | ||
| Ice-box | |||
| Inverted Optic Microscope | Olympus | IX71 | |
| Matrigel Matrix | Becton Dickinson | 354234 | Store at -20 ºC and keep cold when in use. Referred to as basement membrane matrix |
| Methanol | PanReac AppliChem | 131091 | Used for 3D structure fixation prior to immunofluorescent labelling |
| Micropipette | p1000, p200 and p10 | ||
| Microsoft Office Excel | Microsoft | RRID:SCR_016137 | Used to calculate population doubling and to obtain growth rate equation |
| MilliQ water | Referred to as ultrapure water | ||
| Nail Polish | Used to seal samples after mounting | ||
| Parafilm M | Bemis | PM-999 | Used to cover antibody solution during incubation |
| PBS pH 7.4 (w/o calcium & magnesium) | Gibco | 10010-056 | Sterile. Used for cell subculture |
| PBS tablets | Sigma-Aldrich | P4417 | Dilute in milliQ water. No sterility required. Used for immunofluorescence |
| Pipette Aid | |||
| Primaria T25 flasks | Corning | 353808 | Used for BPEC culture |
| Scepter Automated Cell Counter | Millipore | PHCC20060 | Alternatively, use an haemocytometer |
| Scissors | Used to cut pipette tips and parafilm | ||
| Sterile filters 0.22 μm | Millipore | SLGP033RS | Used to filter MTT solution |
| Thiazolyl Blue Tetrazolium Bromide (MTT) | Sigma-Aldrich | M2128 | Store at -20 ºC |
| Triton X-100 | Sigma-Aldrich | T8787 | Used for immunofluorescence of 3D structures |
| Trypsin-EDTA 10X | Biowest | X0930 | Dilute in PBS to obtain 3X solution |
| Vectashield Antifade Mounting Medium | Vector Laboratories | H-1000 | |
| WIT-P-NC Culture Medium | Stemgent | 00-0051 | Used for primary BPEC culture |
| WIT-T Culture Medium | Stemgent | 00-0047 | Used for transformed BPEC culture |
Odniesienia
- Hanahan, D., Weinberg, R. A. Hallmarks of cancer: the next generation. Cell. 144 (5), 646-674 (2011).
- Stampfer, M. R., Yaswen, P. Culture models of human mammary epithelial cell transformation. Journal of Mammary Gland Biology and Neoplasia. 5 (4), 365-378 (2000).
- Schinzel, A. C., Hahn, W. C. Oncogenic transformation and experimental models of human cancer. Frontiers in Bioscience : A Journal and Virtual Library. 13 (13), 71 (2008).
- Balani, S., Nguyen, L. V., Eaves, C. J. Modeling the process of human tumorigenesis. Nature Communications. 8 (1), 15422 (2017).
- Bray, F., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 68 (6), 394-424 (2018).
- Ince, T. A., et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 12 (2), 160-170 (2007).
- Repullés, J., et al. Radiation-induced malignant transformation of preneoplastic and normal breast primary epithelial cells. Molecular Cancer Research. , 1-13 (2019).
- Weigelt, B., Bissell, M. J. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Seminars in Cancer Biology. 18 (5), 311-321 (2008).
- Paoli, P., Giannoni, E., Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochimica et Biophysica Acta. 1833 (12), 3481-3498 (2013).
- Debnath, J., Muthuswamy, S. K., Brugge, J. S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30 (3), 256-268 (2003).
- Borowicz, S., et al. The soft agar colony formation assay. Journal of Visualized Experiments. (92), (2014).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Brocher, J. The BioVoxxel Image Processing and Analysis Toolbox. European BioImage Analysis Symposium. 8 (2), 67112 (2015).
- Torquato, S., Truskett, T. M., Debenedetti, P. G. Is random close packing of spheres well defined. Physical Review Letters. 84 (10), 2064-2067 (2000).
- LaBarge, M. A., Garbe, J. C., Stampfer, M. R. Processing of human reduction mammoplasty and mastectomy tissues for cell culture. Journal of Visualized Experiments. (71), (2013).
- Zubeldia-Plazaola, A., et al. Glucocorticoids promote transition of ductal carcinoma in situ to invasive ductal carcinoma by inducing myoepithelial cell apoptosis. Breast Cancer Research. 20 (1), 65 (2018).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone