A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Methodologies for Studying B. subtilis Biofilms as a Model for Characterizing Small Molecule Biofilm Inhibitors
In This Article
Summary
This study presents the development of reproducible methodologies to study biofilm inhibitors and their effects on Bacillus subtilis multicellularity.
Abstract
This work assesses different methodologies to study the impact of small molecule biofilm inhibitors, such as D-amino acids, on the development and resilience of Bacillus subtilis biofilms. First, methods are presented that select for small molecule inhibitors with biofilm-specific targets in order to separate the effect of the small molecule inhibitors on planktonic growth from their effect on biofilm formation. Next, we focus on how inoculation conditions affect the sensitivity of multicellular, floating B. subtilis cultures to small molecule inhibitors. The results suggest that discrepancies in the reported effects of such inhibitors such as D-amino acids are due to inconsistent pre-culture conditions. Furthermore, a recently developed protocol is described for evaluating the contribution of small molecule treatments towards biofilm resistance to antibacterial substances. Lastly, scanning electron microscopy (SEM) techniques are presented to analyze the three-dimensional spatial arrangement of cells and their surrounding extracellular matrix in a B. subtilis biofilm. SEM facilitates insight into the three-dimensional biofilm architecture and the matrix texture. A combination of the methods described here can greatly assist the study of biofilm development in the presence and absence of biofilm inhibitors, and shed light on the mechanism of action of these inhibitors.
Introduction
Multi-cellular bacterial communities play significant roles in natural and anthropogenic environments, and can be beneficial or highly deleterious. These multi-cellular colonies are known as biofilms, wherein the individual cells are embedded in a self-produced extracellular polymeric substances (EPS) matrix. The EPS strongly adhere the cells to the surface they colonize. They serve as a shield against mechanical and chemical forces and create close contact between neighboring cells, facilitating cellular communication1. A biofilm can be viewed as a differentiated community, where cells use highly regulated, orchestrated processes to coordinate their activities within the community, as well as across species2-5. The transition from a planktonic, free-living mode of growth to a biofilm state is often associated with developmental processes. A good example is the Gram-positive soil bacterium Bacillus subtilis, and therefore an undomesticated strain serves as a robust model organism to study the developmental stages leading to biofilm formation. In this bacterium, motile cells organize themselves into conspicuous multicellular structures that carry out specialized tasks4. One group of cells, the matrix-producers secrete exopolysaccharides6, the amyloid protein TasA7,8, and the surface hydrophobicity protein BslA9,10; all of which participate in the assembly of the EPS11-13.
Given the abundance of biofilms in natural and anthropogenic niches and the putative fatal damage they can cause, there is a pressing need to find ways to prevent their formation. Small molecule inhibitors can aid in the discovery of new regulatory pathways, enzymes and structural proteins involved in biofilm formation, and thereby promote insights in the complex processes of multicellular community assembly. As B. subtilis is a well-studied model for biofilm formation14,15, it can be used to assess the effects of various biofilm inhibitors. This study tackles four fundamental methods that are key for evaluating the response of biofilms to small molecule inhibitors. First, to ensure that these inhibitors have a biofilm-specific target, the separation of the effect on planktonic growth from the effect on biofilm formation is critical. Most antibacterial agents target cells in their planktonic growth phase, but molecules that target the biofilm lifestyle are rare. Additionally, as molecules that do not affect planktonic growth are not toxic, they can reduce the selective pressure favoring antibiotic resistant mutants16. For example, when biofilms are treated with D-amino acids or certain other cell wall-interfering molecules, they are either disturbed or disassembled, but these inhibitors only mildly affect planktonic growth12,17. In contrast, many antibiotics dramatically impair planktonic growth, with little or no effect on biofilm formation17.
Second, establishing a consistent and robust experimental framework to study the effect of the small molecules is crucial. We observed that the active concentration range of small molecule inhibitors was sensitive to the pre-culture conditions and to the experimental setup used to study the effect of these small molecule inhibitors. Various reports, particularly those studying B. subtilis, revealed variations in the concentration range at which D-amino acids inhibit the formation of pellicles — the floating bacterial biofilms12,17-19. The results presented here suggest that the following factors account for differences in the active concentration range: the pre-culture conditions (logarithmic12,17 versus late-stationary20 growth phase), the growth medium used in the pre-culture condition (rich, undefined [Luria Broth, LB] versus defined [monosodium glutamate-glycerol, MSgg]), the inoculation ratio and especially the removal of the pre-culture medium before inoculation. The temperature of static pellicle growth showed a less important role in the activity range of the small molecule inhibitor D-leucine, a representative D-amino acid used in this study.
Finally, once the biofilms are treated with specific biofilm inhibitors, robust and informative methods are required to characterize the effects of these inhibitors on biofilm fitness. Here, two methods to independently characterize the effect of small molecule inhibitors are described in detail: (1) The effect on single cells within a biofilm colony and their resistance to antimicrobial agents. Cells in biofilms are typically more resistant to antibiotics when compared to free-living bacteria21-23. While this phenomenon is multifactorial, the ability of the EPS to reduce antibiotic penetration was often considered as an appealing explanation24. This method assesses the survival of pre-established biofilm cells after exposure to antibacterial substances. (2) The effect on the biofilm colony architecture, from the large to the small scale. Biofilm colonies are characterized by their three-dimensional structure and the presence of the EPS. Utilizing scanning electron microscopy, changes in the cell morphology, biofilm colony structure and the architecture and abundance of the EPS can be visualized from the large (mm) to the small scale (µm).
Protocol
1. Assessing the Effect of Small Molecule Inhibitors on Pellicle and Biofilm Colony Formation
- Prepare a 2x solution of defined biofilm-inducing MSgg medium25 without the calcium chloride and iron(III) chloride hexahydrate. After filter sterilization, add the calcium chloride. The medium is ready to use directly or it can be stored at 4 °C in the dark.
- Prepare the 1x MSgg dilution on the day of the experiment.
- Dilute the 2x MSgg medium to 1x with sterile distilled water (pellicles) or sterile 3% hot (80 °C) agar (biofilms) and add iron(III) chloride hexahydrate to a final concentration of 50 μM (pellicles) or 250 µM (biofilms). Add antibiotics or small molecule inhibitors to the desired concentration and mix well. For example, to obtain a final concentration of 0.5 mM D-leucine in 30 ml to establish pellicles or biofilms, add 196.6 µl of a 76.3 mM (10 mg/ml) D-leucine stock solution.
NOTE: The final 1x MSgg composition is described in Table 1. Compared to the original recipe25, the medium contained 50 µg/ml threonine and the iron concentration to grow biofilm colonies on solid MSgg medium was increased 2.5x to optimize the wrinkled colony morphology.
- Dilute the 2x MSgg medium to 1x with sterile distilled water (pellicles) or sterile 3% hot (80 °C) agar (biofilms) and add iron(III) chloride hexahydrate to a final concentration of 50 μM (pellicles) or 250 µM (biofilms). Add antibiotics or small molecule inhibitors to the desired concentration and mix well. For example, to obtain a final concentration of 0.5 mM D-leucine in 30 ml to establish pellicles or biofilms, add 196.6 µl of a 76.3 mM (10 mg/ml) D-leucine stock solution.
- After solidification of the agar, dry the solid MSgg plates in a biological hood for 30-45 min prior to the inoculation.
- To select specific inhibitors that interfere with the mechanisms of pellicle formation (Figure 1), rule out that the concentrations used affect planktonic and static growth.
- Determine planktonic growth (increase in optical density over time in liquid culture) in a simple growth curve by measuring the optical density at 600 nm every hour until the stationary growth phase.
- To confirm that the measured culture turbidity represents live cell counts, determine the numbers of colony forming units (CFU) of cells in the planktonic growth phase from a shaking culture after several time points.
- To assess the effect of small molecule inhibitors on static pellicle growth, harvest cells at the end of a 3-day incubation at 23 °C from a 24-well cell-culture well, inoculated under the same conditions as described in sections 1.7-1.9 and determine the CFU. For this control, use a pellicle-deficient strain that lacks the operons encoding for the extracellular matrix components (i.e., B. subtilis ΔepsH, ΔtasA).
NOTE: This strain is capable of growing under static conditions, but in contrast to a pellicle-forming wild type, it is deficient in the ability to float to the liquid-air interface, where growth is favored due to increased oxygen levels26. Thus, this extracellular matrix- and pellicle-deficient strain is a recommended reference strain to assess growth under static conditions.
NOTE: For the specific example of the non-canonical D-amino acid D-leucine described below, an effect on planktonic and static growth at concentrations that interfered with pellicle formation was ruled out12,17. The methods to determine planktonic and static growth are described in detail17.
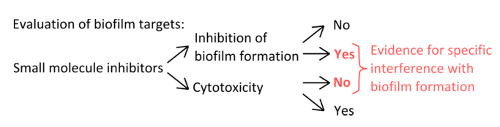
Figure 1. Conceptual overview for the identification of a robust experimental setup to assess the specific inhibition of biofilm formation. Selection criteria for small molecule inhibitors that indicate specific interference with biofilm formation without pronounced effect on planktonic growth. Please click here to view a larger version of this figure.
- Streak out B. subtilis from a -80 °C stock (LB culture of 109 cells/ml frozen in 20% glycerol) to isolate single colonies on a LB-1.5% agar plate with a sterile tip or applicator stick.
- Grow overnight at 30 °C.
- CRITICAL STEP: For a robust pellicle inhibition by the non-canonical D-amino acids such as D-leucine, grow a single colony picked from the LB-1.5% agar plate in 3 ml LB broth at 37 °C for 4 hr in a shaking incubator (shaking speed 200 rpm). Replace the LB broth with biofilm-inducing MSgg medium prior to inoculation by centrifuging 1.5 ml starter culture for 4 min at 6,000 x g, carefully removing the supernatant and re-suspending the pellet in 1.5 ml MSgg medium. The rest of the culture can be discarded.
Important: To ensure the robustness of the system, the optical density at 600 nm (OD600) of the washed starter culture should be between 0.6 and 1. - During the growth of the starter culture, prepare a 12-well cell-culture multidish plate containing 3 ml of MSgg medium without or with a concentration range of the small molecule inhibitor (e.g., 0.3, 0.5, 1 mM D-leucine17). To rule out edge effects, distribute the location of the different concentrations across the multidish plate. Alternatively, use 24-well cell-culture multidish plates containing 1.5 ml of MSgg medium.
- Inoculate the wells of the 12-well cell-culture multidish plate with 3 µl of the washed starter culture (1:1,000 dilution).
NOTE: A lower dilution ratio, i.e., 1:500 can be used. This decreases the development time of the pellicles. - Grow the pellicles at 23 °C under static conditions for three days. Do not move the pellicles during this time, as it can affect the final surface morphology of the pellicle.
- Acquire pictures with a binocular and homogenous exposure of lightning. Alternatively, take a picture of the pellicles with a high resolution camera. To avoid artefacts caused by inconsistent light angles and shadows, take top-down pictures with the camera fixed on a tripod and use a soft and large light source at 45° from both sides.
NOTE: An alternative method to study B. subtilis multicellularity is the biofilm colony assay on solid, biofilm-inducing MSgg medium. Like pellicles, this assay allows the study of spatiotemporal processes. Once the active range of small molecule inhibitors is determined, their effect on biofilm colony formation can be studied. - To grow biofilm colonies, symmetrically spot 1.5 µl of the unwashed pre-culture (Step 1.7) on the dried MSgg 1.5% agar plate with the help of a template — 4 drops per Petri dish of 8.5 cm diameter. Let the drops adsorb to the plate before moving them.
NOTE: The template helps to get an equal distribution of the biofilm colonies within the area where the cells are grown. To prepare the template, draw the total area of growth at original scales, divide it to equal sectors and mark the center. For a round Petri dish of 8.5 cm diameter, this assigns 14 cm2 to one biofilm colony. - Incubate the plates at 30 °C for three days. During this time, biofilm colonies develop and form a three-dimensional, wrinkled structure.
- Take pictures as in step 1.11.
2. Ethanol Resistance Assay
- Grow biofilms as described in steps 1.1-1.7 and 1.12-1.14.
- After 68 hr of growth at 30 °C, cut the biofilm colonies into two equal parts with the help of a razor blade and the template.

Figure 2. Example experimental design to assess the resistance of biofilm colony cells to sterilizing agents. (A) Template used for the equal distribution of biofilm colonies across a Petri dish and for cutting. (B) Top-down images of untreated wild-type biofilm grown for 68 hr on solid, defined biofilm-inducing MSgg medium at 30 °C. The enlargement shows how a biofilm colony can be cut in two equal halves. (C) The two equal biofilm halves are treated equally (control, PBS) or with either PBS or sterilizing agent and processed as described. Scale bar: 1 cm. Please click here to view a larger version of this figure.
- Carefully lift each half of the biofilm colony from the agar plate with a small spatula and move it to a 1.5 ml microcentrifuge tube containing 500 µl of phosphate-buffered saline (PBS). If necessary, scrape the remaining cells from the plate and transfer them to the microcentrifuge tube as well.
NOTE: The second half of the biofilm colony is treated differentially, depending on whether it is the control or to test the resistance to sterilizing agents. - For the control, incubate the second half of the biofilm colony in 500 µl PBS as in step 2.3. To assess resistance to sterilizing agents, transfer the second half of the biofilm colony to 500 µl 50% (v/v) ethanol.
NOTE: Alternative sterilizing agents such as sodium hypochlorite can be used. For all sterilizing agents used, determine the active concentration and incubation time in a preliminary experiment. - Incubate the biofilm colonies for 10 min on the bench-top at room temperature.
- Centrifuge the biofilm colonies for 5 min at 18,000 x g and carefully remove the supernatant with a pipette. Add 300 µl of PBS.
- Sonicate the cells mildly (amplitude 10%, pulse 5 sec) with the microtip of a sonicator.
NOTE: The sonication energy must be sufficient to separate biofilm aggregates. However, too harsh sonication may lyse the cells. Confirm in advance by light microscopy that the sonication energy used does not lyse the cells and that all aggregates are dissolved. - Add 700 µl of PBS to a final volume of 1 ml. Perform a serial dilution (to 10-7) in PBS and spread 100 µl of 3 dilutions on a LB-1.5% agar plate using sterile glass beads.
NOTE: The optimal dilutions to be plated should be determined in a preliminary experiment, as this depends on the amount of cells in the biofilm colony of interest and the survival rate of the cells in response to the sterilizing agent. - Incubate the plates overnight at 30 °C, count the CFU and determine CFU/ml. From the final CFU/ml of each the half biofilm colonies, calculate the percentage of survivors.
NOTE: When performed and analyzed as described, the two halves of the control biofilm colony and the untreated versus ethanol-treated half of the untreated biofilm colony should yield differences below 10% in viable cell counts, verifying the symmetry or the resistance of the colony, respectively. Alternatively, the results can be represented in total CFU. The cell counts of the control and untreated biofilm colony should remain in the same order of magnitude. In contrast, the cell counts of the ethanol-treated half of a small molecule-treated biofilm colony are expected to drop by a minimum of two orders of magnitude to claim for an increased sensitivity to the sterilizing agent.
3. Biofilm Colony Sample Preparation for Scanning Electron Microscopy
- Grow biofilm colonies as described in steps 1.1-1.7 and 1.12-1.14.
- Prepare a fresh batch of 2% (v/v) glutaraldehyde, 3% (v/v) paraformaldehyde solution in 100 mM sodium cacodylate, 5 mM calcium chloride buffer, pH 7.3. Prepare 5 ml of fixative for every Petri dish of 8.5 cm diameter.
CAUTION: Glutaraldehyde and paraformaldehyde are hazardous. Handle them with safety equipment inside a chemical hood. Discard the solutions and the contaminated materials to the hazardous waste. - Carefully add the fixative to the biofilm colonies, without dispensing directly on top of the biofilms.
NOTE: Due the hydrophobic character of the biofilm colony, the colonies slowly detach from the agar and start to float. - Carefully seal the plates with a strip of Parafilm. Incubate on a rotary shaker for 2 hr at room temperature and subsequently transfer the plates to 4 °C for overnight.
- The next day, carefully remove the liquid with a Pasteur glass pipette connected to a vacuum pump.
- Carefully add 10 ml 100 mM sodium cacodylate, 5 mM calcium chloride buffer to wash the biofilm and incubate for 5 min. Gently remove the liquid with the Pasteur glass pipette from the corner of the plate to avoid damaging the biofilm and add fresh washing solution by gentle pipetting. Repeat this step once.
- For the dehydration of the biofilm colonies, proceed with the following steps: 2x 5 min in ddH2O; 2x 20 min in 30% ethanol; 2x 20 min in 50% ethanol; 2x 20 min in 70% ethanol; 2x 20 min in 96% ethanol; 2x 30 min in 100% ethanol.
- Add 15 ml of liquid for every Petri dish of 8.5 cm diameter in each step and remove the liquid carefully after each incubation.
- Use one of two different methods for drying the samples from ethanol.
- For air-drying from ethanol:
- Cut a cellulose filter paper (diameter of 9 cm) in quarters. Briefly submerge one quarter in 100% ethanol, and then carefully transfer one floating biofilm colony onto it. Put the wet filter paper in a Petri dish lined with a filter paper. Cover the Petri dish and let the biofilm colonies dry overnight in a chemical hood.
- For critical point (CP)-drying using carbon dioxide (CO2) as the transition fluid:
- Fill 75% of the critical point drying machine chamber with 100% ethanol. Transfer the samples into a holder, each sample into its own chamber. If necessary, cut the biofilm with scissors into smaller pieces. Leave the samples submerged in ethanol during all the handling. Then, transfer the holder into the chamber and close the chamber tightly.
- Cool the chamber to 7 °C and start stirring. Fill the chamber completely with liquid CO2. During a 7 min incubation time, let the ethanol mix with the CO2. Then, discharge 25% of the solution.
NOTE: Do not empty the chamber below the level of the sample. - Repeat step 3.8.2.2 four times.
- Repeat step 3.8.2.2 five times with an incubation time of 5 min only. Finally, the ethanol should be completely replaced by CO2.
- During the last round, empty only 5% of the chamber. Turn off the stirring and the cooling. Start heating the chamber to 42 °C. At a temperature of 31.1 °C and a pressure of 73.9 bar, the liquid CO2 reaches its critical point, the state where the gaseous phase has the same density as the liquid phase of the solvent27. Once the temperature reaches 42 °C, incubate for 10 min. At 42 °C, the CO2 in the chamber exists as supercritical gas.
NOTE: Constantly check the pressure of the chamber. The pressure should not exceed 120 bar at 42 °C. - Start to slowly release the gas with continuous heating. This keeps the samples in the CO2-gas phase and prevents the deformation of the sample morphology through the liquid surface tension. Set the flowmeter to 5 L/hr by fine tuning the metering valve controlling the flowmeter. Wait until the all the pressure in the chamber is released. Now open the chamber and remove the samples carefully from the holder.
- For air-drying from ethanol:
- Coat an electron microscopy stub with carbon tape. With the help of tweezers, carefully transfer the biofilm colonies onto the stub. Connect each colony to the stub by adding a thin bridge from carbon tape, which is crucial for charge elimination under the electron beam. At this stage, handle the biofilm colonies with care as they are very fragile. Store the samples in a desiccator for at least 24 hr or until examination.
- The day of the examination with the scanning electron microscope, sputter-coat the biofilm colonies for 2 min in a 60° angle in a gold-palladium sputter-coater. Repeat this step twice and rotate the samples by 120° in between. At the end, sputter-coat the samples once for 3 min from the top. The 20 nm thin layer of gold-palladium improves the conductivity and enhances the contrast of the sample for the imaging in the SEM.
- Store the samples in a desiccator to avoid the rehydration of the sample28 until imaging with a scanning electron microscope29,30.
Results
The pellicle assay is one method to study the highly regulated and dynamic processes of B. subtilis multicellularity. Besides this, the pellicle assay is suited to test a range of either pre-starter conditions or small molecule concentrations in a single cell-culture multidish plate in one experiment. However, B. subtilis pellicle formation is sensitive to the pre-culture conditions (e.g., growth medium of the pre-culture and its growth phase), the inoculation ratio and the removal of the pre-c...
Discussion
Bacillus subtilis forms robust and highly structured biofilms both in liquid (pellicles) and on solid medium (colonies). Hence, it serves as an ideal model organism to characterize the mode of action of specific biofilm inhibitors. On solid media, cells form multicellular structures with distinctive features that are not evident in pellicles, like wrinkles radiating from the center to the edge. Thus, pellicles and colonies are complementary systems to study B. subtilis multicellularity.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Electron microscope imaging was conducted at the Electron Microscopy Unit of the Weizmann Institute of Science, supported in part by the Irving and Cherna Moskowitz Center for Nano and Bio-Nano Imaging. This research was also supported by the ISF I-CORE grant 152/1, Mr. and Mrs. Dan Kane, Ms. Lois Rosen, by a Yeda-Sela research grant, by the Larson Charitable Foundation, by Ruth and Herman Albert Scholars Program for New Scientists, by the Ilse Katz Institute for Materials Sciences and Magnetic Resonance Research grant, by the Ministry of Health grant for alternative research methods, and by the France-Israel Cooperation - Maimonide-Israel Research Program. IKG is a recipient of the Rowland and Sylvia Career Development Chair.
Materials
| Name | Company | Catalog Number | Comments |
| Luria Broth, Lennox | Difco | 240230 | |
| Bacto Agar | Difco | 214010 | |
| potassium phosphate monobasic | Sigma, 136.09 g/mol | P0662-500G | |
| potassium phosphate dibasic | Fisher Scientific, 174.18 g/mol | BP363-1 | |
| 3-(N-morpholino)propanesulfonic acid | Fisher Scientific, 209.27 g/mol | BP308-500 | |
| magnesium chloride hexahydrate | Merck, 203.30 g/mol | 1.05833.0250 | |
| calcium chloride anhydrous | J.T. Baker, 110.98 g/mol | 1311-01 | |
| manganese(II) chloride tetrahydrate | Sigma, 197.91 g/mol | 31422-250G-R | |
| iron(III) chloride hexahydrate | Sigma, 270.30 g/mo) | F2877-500G | |
| zinc chloride anhydrous | Acros Organics, 136.29 g/mol | 424592500 | |
| thiamine hydrochloride | Sigma, 337.27 g/mol | T1270-100G | |
| L-tryptophan | Fisher Scientific, 204.1 g/mol | BP395-100 | |
| L-phenylalanine | Sigma, 165.19 g/mol | P5482-100G | |
| L-threonine | Sigma, 119.12 g/mol | T8625-100G | |
| glycerol anhydrous | Bio-Lab Itd | 712022300 | |
| L-glutamic acid monosodium salts hydrate | Sigma, 169.11 g/mol | G1626-1KG | |
| D-leucine | Sigma, 169.11 g/mol | 855448-10G | |
| ethanol anhydrous | Gadot | 830000054 | |
| razor blade | Eddison | NA | |
| circular cellulose filter papers | Whatman, 90 mm | 1001-090 | |
| glutaraldehyde | EMS (Electron Micoscopy Science), 25% in water | 16220 | |
| paraformaldehyde | EMS, 16% in water | 15710 | |
| sodium cacodylate | Merck, 214.05 g/mol | 8.2067 | |
| calcium chloride 2-hydrate | Merck, 147.02 g/mol | 1172113 | |
| stub-aluminium mount | EMS, sloted head | 75230 | |
| carbon adhesive tape | EMS | 77825-12 | |
| Shaker 37 °C | New Brunswick Scientific Innowa42 | NA | |
| Centrifuge | Eppendorf table top centrifuge 5424 | NA | |
| Digital Sonifier, Model 250, used with Double Step Microtip | Branson | NA | |
| Incubator 30 °C | Binder | NA | |
| Incubator 23 °C | Binder | NA | |
| Filter System, 500 ml, polystyrene | Cornig Incorporated | NA | |
| Rotary Shaker - Orbitron Rotatory II | Boekel | NA | |
| S150 Sputter Coater | Edwards | NA | |
| CPD 030 Critical Point Dryer | BAL-TEC | NA | |
| Environmental Scanning Electron Microscope | XL30 ESEM FEG Philips (FEI) | NA |
References
- Branda, S. S., Vik, S., Friedman, L., Kolter, R. Biofilms: the matrix revisited. Trends Microbiol. 13, 20-26 (2005).
- Stoodley, P., Sauer, K., Davies, D. G., Costerton, J. W. Biofilms as complex differentiated communities. Annu Rev Microbiol. 56, 187-209 (2002).
- Miller, M. B., Bassler, B. L. Quorum sensing in bacteria. Annu Rev Microbiol. 55, 165-199 (2001).
- Aguilar, C., Vlamakis, H., Losick, R., Kolter, R. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol. 10, 638-643 (2007).
- Kolter, R., Greenberg, E. P. Microbial sciences: the superficial life of microbes. Nature. 441, 300-302 (2006).
- Kearns, D. B., Chu, F., Branda, S. S., Kolter, R., Losick, R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 55, 739-749 (2005).
- Branda, S. S., Chu, F., Kearns, D. B., Losick, R., Kolter, R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 59, 1229-1238 (2006).
- Romero, D., Aguilar, C., Losick, R., Kolter, R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci USA. 107, 2230-2234 (2010).
- Kobayashi, K., Iwano, M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 85, 51-66 (2012).
- Hobley, L., et al. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci USA. 110, 13600-13605 (2013).
- Romero, D., Vlamakis, H., Losick, R., Kolter, R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol. 80, 1155-1168 (2011).
- Kolodkin-Gal, I., et al. D-amino acids trigger biofilm disassembly. Science. 328, 627-629 (2010).
- Chan, Y. G., Kim, H. K., Schneewind, O., Missiakas, D. The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J Biol Chem. 289, 15680-15690 (2014).
- Mielich-Suss, B., Lopez, D. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol. 17, 555-565 (2014).
- Cairns, L. S., Hobley, L., Stanley-Wall, N. R. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol Microbiol. 93, 587-598 (2014).
- Chen, M., Yu, Q., Sun, H. Novel strategies for the prevention and treatment of biofilm related infections. Int J Mol Sci. 14, 18488-18501 (2013).
- Bucher, T., Oppenheimer-Shaanan, Y., Savidor, A., Bloom-Ackermann, Z., Kolodkin-Gal, I. Disturbance of the bacterial cell wall specifically interferes with biofilm formation. Environ Microbiol Rep. 7, 990-1004 (2015).
- Sarkar, S., Pires, M. M. D-Amino acids do not inhibit biofilm formation in Staphylococcus aureus. PLoS One. 10, e0117613 (2015).
- Wei, W., Bing, W., Ren, J., Qu, X. Near infrared-caged D-amino acids multifunctional assembly for simultaneously eradicating biofilms and bacteria. Chem Commun (Camb). 51, 12677-12679 (2015).
- Leiman, S. A., et al. D-amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. J Bacteriol. 195, 5391-5395 (2013).
- Costerton, J. W., Stewart, P. S., Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science. 284, 1318-1322 (1999).
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2, 114-122 (2003).
- Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis. 34, 877-886 (2015).
- Tseng, B. S., et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 15, 2865-2878 (2013).
- Branda, S. S., Gonzalez-Pastor, J. E., Ben-Yehuda, S., Losick, R., Kolter, R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 98, 11621-11626 (2001).
- Holscher, T., et al. Motility, Chemotaxis and Aerotaxis Contribute to Competitiveness during Bacterial Pellicle Biofilm Development. J Mol Biol. 427, 3695-3708 (2015).
- Bray, D. . Methods in Biotechnology. 13, 235-243 (2000).
- Ensikat, H. J., Ditsche-Kuru, P., Barthlott, W. . Scanning electron microscopy of plant surfaces: simple but sophisticated methods for preparation and examination. 1, 248-255 (2010).
- Hayat, M. A. . Principles and techniques of scanning electron microscopy: Biological applications. 2, (1976).
- Schatten, H. . Scanning Electron Microscopy for the Life Sciences. , (2013).
- Bridier, A., Meylheuc, T., Briandet, R. Realistic representation of Bacillus subtilis biofilms architecture using combined microscopy (CLSM, ESEM and FESEM). Micron. 48, 65-69 (2013).
- Boyde, A., MacOnnachie, E. Volume changes during preparation of mouse embryonic tissue for scanning electron microscopy. SCANNING. 2, 149-163 (1979).
- Yao, Z., Kahne, D., Kishony, R. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol Cell. 48, 705-712 (2012).
- Epstein, A. K., Pokroy, B., Seminara, A., Aizenberg, J. Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc Natl Acad Sci USA. 108, 995-1000 (2011).
- Vlamakis, H., Chai, Y., Beauregard, P., Losick, R., Kolter, R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 11, 157-168 (2013).
- Shemesh, M., Chai, Y. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J Bacteriol. 195, 2747-2754 (2013).
- Kolodkin-Gal, I., et al. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 27, 887-899 (2013).
- Oppenheimer-Shaanan, Y., et al. Spatio-temporal assembly of functional mineral scaffolds within microbial biofilms. npj Biofilms and Microbiomes. 2, 15031 (2016).
- Garcia-Betancur, J. C., Yepes, A., Schneider, J., Lopez, D. Single-cell analysis of Bacillus subtilis biofilms using fluorescence microscopy and flow cytometry. J Vis Exp. , e3796 (2012).
- Bogino, P. C., Oliva Mde, L., Sorroche, F. G., Giordano, W. The role of bacterial biofilms and surface components in plant-bacterial associations. Int J Mol Sci. 14, 15838-15859 (2013).
- Fratamico, P. M., Annous, B. A., Guenther, N. W. . Biofilms in the Food and Beverage Industires. 1, (2009).
- Gao, G., et al. Effect of biocontrol agent Pseudomonas fluorescens 2P24 on soil fungal community in cucumber rhizosphere using T-RFLP and DGGE. PLoS One. 7, e31806 (2012).
- Chen, Y., et al. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol. 15, 848-864 (2013).
- Bryers, J. D. Medical biofilms. Biotechnol Bioeng. 100, 1-18 (2008).
- Logan, B. E. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol. 7, 375-381 (2009).
- Nevin, K. P., Woodard, T. L., Franks, A. E., Summers, Z. M., Lovley, D. R. Microbial electrosynthesis: feeding microbes electricity to convert carbon dioxide and water to multicarbon extracellular organic compounds. MBio. 1, (2010).
- Torres, C. I., et al. A kinetic perspective on extracellular electron transfer by anode-respiring bacteria. FEMS Microbiol Rev. 34, 3-17 (2010).
- Li, J., Wang, N. Foliar application of biofilm formation-inhibiting compounds enhances control of citrus canker caused by Xanthomonas citri subsp. citri. Phytopathology. 104, 134-142 (2014).
- Okegbe, C., Price-Whelan, A., Dietrich, L. E. Redox-driven regulation of microbial community morphogenesis. Curr Opin Microbiol. 18, 39-45 (2014).
- Mann, E. E., Wozniak, D. J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 36, 893-916 (2012).
- Bouffartigues, E., et al. Sucrose favors Pseudomonas aeruginosa pellicle production through the extracytoplasmic function sigma factor SigX. FEMS Microbiol Lett. 356, 193-200 (2014).
- Wu, C., Lim, J. Y., Fuller, G. G., Cegelski, L. Quantitative analysis of amyloid-integrated biofilms formed by uropathogenic Escherichia coli at the air-liquid interface. Biophys J. 103, 464-471 (2012).
- Serra, D. O., Richter, A. M., Hengge, R. Cellulose as an Architectural Element in Spatially Structured Escherichia coli Biofilms. J Bacteriol. 195, 5540-5554 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved