A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Quantitative Analysis of Cell Edge Dynamics during Cell Spreading
* These authors contributed equally
In This Article
Summary
In this protocol, we present the experimental procedures of a cell spreading assay that is based on live-cell microscopy. We provide an open-source computational tool for the unbiased segmentation of fluorescently labeled cells and quantitative analysis of lamellipodia dynamics during cell spreading.
Abstract
Cell spreading is a dynamic process in which a cell suspended in media attaches to a substrate and flattens itself from a rounded to a thin and spread-out shape. Following the cell-substrate attachment, the cell forms a thin sheet of lamellipodia emanating from the cell body. In the lamellipodia, globular actin (G-actin) monomers polymerize into a dense filamentous actin (F-actin) meshwork that pushes against the plasma membrane, thereby providing the mechanical forces required for the cell to spread. Notably, the molecular players that control the actin polymerization in lamellipodia are essential for many other cellular processes, such as cell migration and endocytosis.
Since spreading cells form continuous lamellipodia that span the entire cell periphery and persistently expand outward, cell spreading assays have become an efficient tool to assess the kinetics of lamellipodial protrusions. Although several technical implementations of the cell spreading assay have been developed, a detailed description of the workflow, which would include both a step-by-step protocol and computational tools for data analysis, is currently lacking. Here, we describe the experimental procedures of the cell spreading assay and present an open-source tool for quantitative and unbiased analysis of cell edge dynamics during spreading. When combined with pharmacological manipulations and/or gene-silencing techniques, this protocol is amenable to a large-scale screen of molecular players regulating lamellipodial protrusions.
Introduction
Lamellipodial protrusions are prominent cytoskeletal structures formed at the front of a migrating cell. In lamellipodia, polymerization of actin with the aid of the Arp2/3 complex and formins creates a fast-growing branched actin meshwork that pushes against the plasma membrane1,2. The pushing force generated by the actin meshwork physically propels the cell forward1,3,4,5. Depletion of the Arp2/3 complex or disruption of signaling pathways essential for lamellipodial protrusions often impair cell migration6, 7. Although migration of lamellipodia-deficient cells has also been reported8,9, the importance of lamellipodia in cell migration is evident as depletion of this protrusive structure perturbs the cell's ability to move through complex biological microenvironments6,10.
A major hindrance to understanding the regulation of lamellipodia in migrating cells is the natural variability in lamellipodial protrusion kinetics, size, and shape11,12,13,14. Furthermore, recent studies have demonstrated that lamellipodia exhibit complex protrusive behaviors, including fluctuating, periodic, and accelerating protrusions14,15. Compared to the highly variable lamellipodia of migrating cells6,16, lamellipodia formed during cell spreading are more uniform12. Since the protrusive activity of spreading and migrating cells is driven by identical macromolecular assemblies, which include a branched actin network, contractile actomyosin bundles, and integrin-based cell-matrix adhesions17,18, spreading cells have been widely used as a model for investigating the regulation of lamellipodia dynamics.
Cell spreading is a dynamic mechanochemical process where a cell in suspension first adheres to a substrate through integrin-based adhesions17,19,20 and then spreads by extending actin-based protrusions21,22,23. During the spreading phase, lamellipodia emanating from the cell body protrude isotropically and persistently with little to no retraction or stalling12. The most commonly used cell spreading protocols are endpoints assays, where spreading cells are fixed at various times after plating19,24. These assays, although quick and simple, are limited in their diagnostic power to detect changes in the dynamic features of lamellipodia. To determine the molecular mechanisms that control lamellipodia dynamics, the Sheetz group pioneered the use of quantitative analysis of live spreading cells and uncovered many fundamental properties of cell edge protrusions11,12,22. These studies have demonstrated that the live-cell spreading assay is a robust and powerful technique in the toolbox of a cell biology laboratory. Despite that, a detailed protocol and open-source computational tool for a live-cell spreading assay are currently unavailable for the cell biology community. To this end, our protocol outlines the procedures of imaging live spreading cells and provides an automated image analysis tool. To validate this method, we used Arp2/3 inhibition as an experimental treatment and showed that inhibiting the function of the Arp2/3 complex did not arrest cell spreading but caused a significant reduction in cell protrusion speed, as well as the stability of cell edge protrusions, giving rise to jagged cell edges. These data demonstrate that the combination of live-cell imaging and automated image analysis is a useful tool for analyzing cell edge dynamics and identifying molecular components that regulate lamellipodia.
Protocol
1. Cell Seeding
NOTE: The described cell spreading protocol was performed using mouse embryonic fibroblasts (MEFs) expressing PH-Akt-GFP (a fluorescent marker for PIP3/PI(3,4)P2). This cell line was generated by genomically integrating an expression construct for PH-Akt-GFP (Addgene #21218) by CRISPR-mediated gene editing. However, other fluorescent markers that are expressed transiently or integrated in the genome can also be used in this assay. For optimal image segmentation, we recommend using fluorescent markers that are evenly distributed in the cytoplasm, e.g., cytosolic GFP.
- Culture a 10 cm dish of cells to 90% confluency.
- Once the cells have attained the proper confluency, place a 22 mm x 22 mm coverslip (#1.5; 0.17 mm thickness) into a 35 mm cell culture dish. Coat the coverslip with 400 µL of fibronectin that has been diluted in PBS to a final concentration of 2.5 µg/mL.
NOTE: The number of coverslips required for the assay is determined by the number of experimental conditions and technical replica. - Place the 35 mm dish with the fibronectin-coated coverslip into a 37 °C, 5% CO2 incubator for 1 hour.
- Remove the dish with the coverslip from the incubator. Aspirate the fibronectin and wash the coverslip with PBS by gently pipetting around the coverslip two to three times.
- Aspirate the cell culture media from the 10 cm dish of cells and wash the dish with PBS.
- Add 650 µL of 0.05% trypsin-EDTA to the 90% confluent dish of cells, tilting the dish to evenly distribute the enzyme. Place the dish with the trypsin into the incubator for 1 minute.
- Remove the dish with the cells from the incubator. Add 10 mL of cell culture media into a 15 mL centrifuge tube. Quickly add another 10 mL of media into the dish to quench the trypsin.
- Pipette 1 mL of the trypsinized cells into the 15 mL centrifuge tube in order to dilute the cells. Pipette the contents of the tube up and down to ensure an even distribution of cells within the media. For cell types with high aggregation propensity, filtering cells through a cell strainer (100 µm mesh size) is recommended to minimize the occurrence of cell clumping.
- From the tube, pipette 500 - 1000 µL of diluted cells into the 35 mm dish containing the coverslip.
- Gently shake the dish to evenly spread out the cells. Ensure that the coverslip is at a ~10% confluency (~50,000 cells/mL) and adjust the volume of diluted cells as needed.
NOTE: The purpose of having cells at such low confluency is to ensure that there are 1-2 polarized cells in each field of view that will be used to focus the objective throughout the cell spreading acquisition. - Passage 1/5 of the remaining cells in the 10 cm dish into one 6 cm dish per treatment condition. Place the passaged dishes and the 35 mm dish with the coverslip into the incubator overnight.
NOTE: These will be the cells that will be analyzed for spreading dynamics.
2. Drug Incubation and Cell Recovery
- Add 5 mL of cell culture media into each of two 15 mL centrifuge tubes, and 20 mL of phenol red free DMEM into each of two 50 mL centrifuge tubes.
NOTE: The number of tube pairs (15 mL + 50 mL) should correspond to the number of experimental conditions. - To test the importance of Arp2/3 for cell spreading, pipette either the pharmacological inhibitor of Arp2/3, CK-666, or the control treatment, such as DMSO, into each pair of centrifuge tubes up to the desired concentration.
- Remove the passaged 6 cm dishes (see Step 1.11) from the incubator and aspirate the media. Wash the dishes with warm PBS.
- Add the contents of the CK-666- or DMSO-supplemented 15 mL centrifuge tubes into each of the passaged dishes. Label each dish with the correct drug treatment and place the dishes into the incubator for one hour.
- Remove the dishes from the incubator and aspirate the media. Wash the dishes with warm PBS in order to thoroughly remove all the remaining phenol red media.
- Add 230 µL of 0.05% trypsin-EDTA to each 6 cm dish and incubate cells for 1 minute.
NOTE: If applicable, trypsin can be replaced by a non-proteolytic cell adhesion blocker. - Remove the dishes from the incubator. For each treatment, add 5 mL of drug-supplemented phenol red free DMEM into a 15 mL centrifuge tube designated as "Tube B". Add an additional 5 mL of the same media into the relevant dish to quench the trypsin. Transfer the contents of the dish into a 15 mL centrifuge tube designated as "Tube A".
- Transfer 1 mL of cells from Tube A into Tube B. Repeat for each treatment.
- Place Tubes A and B into the incubator for 45 minutes to allow cells to recover from trypsinization. Slightly loosen the cap of the centrifuge tubes before placing them in the incubator to allow for CO2 penetrance.
NOTE: The duration of recovery time may vary for different cell types. Although in our experiments 45-minute-long recovery had a negligible effect on cell viability, some cell types may undergo anoikis when maintained in suspension for too long. Therefore, we recommend determining the optimal recovery time empirically. The optimal recovery time enables fast and synchronous cell spreading with no dead or apoptotic cells in the sample.
3. Magnetic Chamber Preparation
- Ensure that all parts of a 1 Well Chamlide Cell Magnetic Chamber that can accommodate a 22 mm x 22 mm square coverslip have been cleaned before use.
- Remove the 35 mm dish with the coverslip (see Step 1.11) from the incubator. Aspirate the cell culture media and wash the coverslip with warm PBS.
- Remove the coverslip from the 35 mm dish using a pair of forceps and gently lay the coverslip onto the bottom plate of the magnetic chamber.
- Place the silicone gasket on top of the coverslip.
NOTE: An improperly placed silicone gasket is the most common cause of a leaky magnetic chamber. Ensure that the gasket rests in the indent of the bottom plate and does not rise beyond the indent. - Attach the main body onto the bottom plate.
NOTE: Do this part very slowly. A good tip is to hold down the bottom plate with one hand while placing the main body on top. This ensures that the main body's magnets do not lift the bottom plate up, which could potentially displace and crack the coverslip. - Add 1 mL of drug-supplemented phenol red free DMEM to the magnetic chamber. Take a lint-free tissue and carefully dab the enclosure between the main body and the bottom plate in order to check for any leaks.
NOTE: If there is leakage, quickly aspirate the media and proceed again from step 3.4. - Lower the transparent cover onto the main body to enclose the magnetic chamber.
- Spray a laboratory tissue first with water and wipe the bottom of the magnetic chamber (the coverslip, not the metal part). Afterwards, spray a second laboratory tissue with a small amount of 70% ethanol and wipe, being careful not to crack the coverslip.
4. Image Acquisition
- Preheat the stage top incubator and the objective heater to 37 °C and set the CO2 level in the stage top incubator to 5%.
NOTE: If the stage top incubator is not connected to a CO2 supply, the cell culture media should be supplemented with 25 mM HEPES to maintain constant pH 7.4. - Apply a sufficient amount of immersion oil to the pre-warmed 60X, 1.4 N.A. oil objective.
NOTE: We use a 60X, 1.4 N.A. oil immersion objective in this protocol because of its reasonably large field of view and outstanding light collection efficiency. If a larger field of view is required, a lower magnification objective (e.g., 20x) can be used as long as the signal-to-noise ratio of the images is greater than 2.5. - Bring both the completed magnetic chamber and Tube B (Step 2.9) to the confocal microscope. Place the magnetic chamber onto the stage top incubator.
NOTE: Place the magnetic chamber gently on the stage to avoid creating bubbles in the immersion oil. - Set focus to the fluorescent cells using the GFP channel. Make sure that the cell edge is sharp and well defined.
- Remove the transparent cover of the magnetic chamber and pipette 500 µL from Tube B into the magnetic chamber. Place the transparent cover back on top of the magnetic chamber.
- To identify cells ideal for cell spreading analysis, search for "halos" of cells that have yet to attach to the coverslip but are no longer rolling around. Cells that are in the earliest stages of coverslip attachment are also great candidates, but image acquisition must be swift in order to capture spreading.
- Configure the time-lapse image acquisition for the green channel to include four fields of view, imaged at 6 second intervals.
NOTE: Due to the high variability of lamellipodia protrusion velocity among different cell types, the optimal frame rate should be determined empirically. The imaging interval of 6 seconds used in our experiments is a good starting point for the analysis of many mesenchymal and epithelial cells. However, cells that spread very quickly (e.g., immune cells) may require a much higher frame rate (shorter imaging interval). The optimal frame rate for cell spreading movies ensures a 2-5 pixels displacement of the protruding cell edge between subsequent frames. Considering the accuracy of curve fitting used to identify the plateau of cell spreading, the optimal frame rate should also ensure 50-100 measurements of cell edge displacement during the rapid expansion phase of cell spreading. The number of fields of view should be adjusted depending on the exposure time, the distance between acquisition points, and the stage movement speed. Users are advised to determine the maximum number of fields of view that can acquired with the desired frame rate. - After identifying a suitable field of view, save the X and Y coordinates of the microscope stage. Proceed with identifying three other fields of view that are relatively close to one another on the coverslip. Save the coordinates of the microscope stage for every desired field of view.
NOTE: It is strongly recommended to optimize the stage movement path between fields of view in order to minimize any unnecessary sample movement. Such optimization can be performed either manually or automatically. Excessive sample movement slows down the acquisition and may cause cells to roll out of view as they are descending. - Acquire images for 15 minutes at a 6 second frame rate and save the files. If more acquisitions are required, repeat starting at Step 4.6.
5. Analysis of cell area, circularity and protrusion dynamics during cell spreading
- Prepare images for data processing and analysis
NOTE: The software requires an image in .tiff format and a pixel size as the input parameters. Both requirements can be fulfilled using the acquisition software or Fiji (in this protocol). If these requirements are fulfilled, proceed to step 5.2.- Install the latest version of Fiji application (https://imagej.net/Fiji/Downloads).
- Open a time-lapse image using Fiji.
- Copy the pixel size of the image by selecting Image > Properties. Copy and paste the pixel size in µm to Notepad/Word.
- For the analysis of cell spread area and circularity, save the time-lapse image as a tiff image stack. The custom-build analysis software does not support proprietary file formats. Save the individual cell tiff image stack by selecting File > Save As > Tiff.
- Install the Python IDE (Spyder) and the necessary packages (PySimpleGUI and tifffile) for data processing and analysis.
NOTE: The installation of Python and packages is only required for the initial setup.- The time-lapse movies will be analyzed in the Spyder IDE using a custom-build Python script. To download the Spyder IDE, download the Anaconda distributor (https://www.anaconda.com/products/individual) which includes Spyder IDE and most of the necessary libraries and packages for this analysis.
- Install Anaconda and launch Spyder through Anaconda Navigator.
- In the IPython console tab (located in the lower right section of Spyder), copy and paste the following command: pip install PySimpleGUI and press the enter key. Running this command will install the package needed to initiate the graphical user interface (GUI).
- In the same console, copy and paste the following command: pip install tifffile and press the Enter key. Running this command will install the package needed to save images as tiff files.
- Download all the Python scripts from the supplemental files or the most updated scripts from GitHub at: https://github.com/ernestiu/Cell-spreading-analysis.git
- Quantify cell area and cell shape factors during cell spreading
- Open the main analysis script "cell_spreading_GUI.py" by selecting the open file option in the top panel of Spyder or using the shortcut Ctrl + O.
- Open the cell spreading analysis GUI by selecting "Run file" in the top panel or using the shortcut F5.
- Click the Cell Spread Area tab (Figure 3A).
- Select the tiff image to be analyzed using the browse button.
NOTE: The selected file must be a tiff file. - Specify the destination directory where data outputs (e.g., cell masks, values) will be saved.
- Specify the data output settings:
- Save masks: Save cell masks generated during the segmentation process.
- Export data: Export an excel spreadsheet (.xlsx) that contains all the analysis data to the destination folder.
Cell area, circularity and aspect ratio of all spreading cells will be saved as an Excel spreadsheet in the destination folder. The cell area is calculated as:
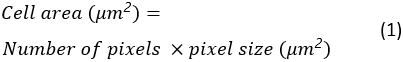
The cell circularity is a measure of how close a cell is to a perfectly round cell. It is calculated as:
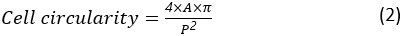
where A and P are the cell area and the cell perimeter, respectively. The aspect ratio of the cell represents how elongated the cell is. A spreading cell should have an aspect ratio close to 1. The aspect ratio is calculated as:
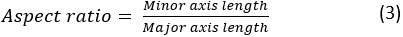
- Save contours: Save the cell boundary contour overlay images in the destination folder.
- Specify the segmentation settings
- Show segmentation: Show the segmentation result in the Spyder console during the analysis process.
- Smallest cell area (µm2): Enter the minimum value for cell area, including area values of cells in the beginning stages of attachment. Objects with an area smaller than this threshold will not be considered as spreading cells. This number will affect the segmentation process.
- Specify the image parameters.
- Acquisition interval (s): Enter the frequency of the image acquisition in seconds.
- Pixel size (µm): Enter the pixel size that was recorded when preparing images for analysis.
- Image bit depth: Enter the bit depth of the camera/detector.
- Click Run. If an error arises, an error message will appear in Spyder's console. Otherwise, the image analysis process will be shown in the console.
NOTE: The first image to appear in the console/Plots section (depending on the Spyder settings) shows all the cells identified in the field of view. Green boxes placed around the cells indicate spreading cells that are suitable for segmentation and analysis. Grey boxes indicate cells that are not suitable for analysis. The total number of identified spreading cells will also appear in the Console tab. The software plots the cell area (in blue) and cell circularity (in red) as a function of time. These graphs allow users to evaluate the accuracy of cell segmentation. A successful segmentation yields a monotonically increasing curve for cell area. To obtain a representative curve of cell spread area, the lag phase should be removed from the graph manually. The lag phase includes measurements of cell area before the cell starts spreading. The lag phase is indicated by fast fluctuations, as represented in the cell area plot (Figure 3C right).
6. Quantify cell edge dynamics during cell spreading using kymographs
- Prior to running the analysis, crop the raw movies of spreading cells to create time series of individual spreading cells.
- Use the Rectangle tool in the Fiji tool bar to manually select a region of interest (ROI) that encapsulates a single cell. (To ensure that the ROI fully encapsulates the spreading cell, use the scroll function to inspect the ROI at all time points.)
- Right click on the ROI and select Duplicate.
- Check Duplicate stack and click OK.
- Open the main analysis script "cell_spreading_GUI.py" by selecting the Open File button in Spyder's tool bar or using the shortcut Ctrl + O. If the GUI has already been opened, go directly to Step 6.3.
- Open the cell spreading analysis GUI by selecting Run file in the top panel or using the shortcut F5 (Figure 3B).
- Click the Kymograph generator & analysis tab.
- Use the browse button to select the tiff image for the analysis.
NOTE: Proprietary file formats, e.g., nd2, lif, zen, are not supported by the script. - Specify the destination folder to save the output data (cell masks and values).
- Specify the output settings.
- Export data: Export an excel spreadsheet (.xlsx) to the destination folder that contains relative cell edge positions and retraction events of the kymographs.
- Specify the image parameters:
- Acquisition interval (s): Enter the image acquisition frequency in seconds.
- Pixel size (µm): Enter the pixel size that was recorded when preparing images for the analysis in Step 5.1.3.
- Smallest cell area (µm2): Enter the minimum value for cell area, including area values of cells in the beginning stages of attachment. Objects with an area smaller than this threshold will not be considered as spreading cells. This number will affect the segmentation process.
- Image bit depth: Enter the bit depth of the camera/detector.
- Click Run. If an error arises, an error message will appear in Spyder's console. Otherwise, a summary of the protrusion dynamics quantifications will be shown in the console. There will be 4 pairs of retraction frequency and protrusion speed measurements, which are extracted from 4 kymographs generated from the top, bottom, left and right portions of the cell. The retraction frequency is calculated as:
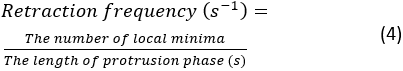
NOTE: This number demonstrates how frequently the lamellipodium retracts over the course of spreading. The average protrusion speed is measured by the slope between the beginning of protrusion and the plateau point on the kymograph. A summary kymograph figure will be shown in the console after the segmentation. To save the summary figure, right click on the figure and save the image.
Results
The above protocol describes the experimental procedures for the live-cell imaging of spreading cells and a computational tool for the quantitative analysis of cell spreading dynamics. The computational tool can be used in a low- or high-throughput format to identify the molecular players regulating the actin polymerization machinery at the cell leading edge.
The schematic representation of the experimental procedures is depicted in Figure 1. The cell spreading as...
Discussion
The described cell spreading assay allows for the continuous tracking of morphological changes (e.g., cell size and shape) and cell edge movements (i.e., protrusion speed and retraction frequency), which are features missing in most cell spreading protocols19,24. While commonly used end-point cell spreading assays allow for the determination of cell spreading speed, these assays fail to resolve the temporal dynamics of cell edge movements. The l...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Connaught Fund New Investigator Award to S.P., Canada Foundation for Innovation, NSERC Discovery Grant Program (grants RGPIN-2015-05114 and RGPIN-2020-05881), University of Manchester and University of Toronto Joint Research Fund, and University of Toronto XSeed Program.
Materials
| Name | Company | Catalog Number | Comments |
| 0.05% Trypsin (0.05%), 0.53 mM EDTA | Wisent Bioproducts | 325-042-CL | |
| 10.0 cm Petri Dish, Polystyrene, TC Treated, Vented | Starstedt | 83.3902 | |
| 15 mL High Clarity PP Centrifuge Tube, Conical Bottom, with Dome Seal Screw Cap, Sterile | Falcon | 352097 | |
| 1-Well Chamlide CMS for 22 mm x 22 mm Coverslip | Quorum Technologies | CM-S22-1 | |
| 35 mm TC-treated Easy-Grip Style Cell Culture Dish | Falcon | 353001 | |
| 50 mL Centrifuge Tube, Transparent, Plug Seal | Nest | 602002 | |
| 6.0 cm Cell Culture Dishes Treated for Increased Cell Attachment, Sterile | VWR | 10861-658 | |
| Arp2/3 Complex Inhibitor I, CK-666 | Millipore Sigma | 182515 | |
| Camera, Prime 95B-25MM | Photometrics | ||
| Dimethyl Sulfoxide, Sterile | BioShop | DMS666 | |
| DMEM, 1x, 4.5 g/L Glucose, with L-Glutamine, Sodium Pyruvate and Phenol Red | Wisent Bioproducts | 319-005 CL | |
| DMEM/F-12, HEPES, No Phenol Red | Gibco | 11039021 | |
| D-PBS, 1X | Wisent Bioproducts | 311-425 CL | |
| Fetal Bovine Serum | Wisent Bioproducts | 080-110 | |
| Fiji Software | ImageJ | ||
| HEPES (1 M) | Gibco | 15630080 | |
| Human Plasma Fibronectin Purified Protein 1 mg | Millipore Sigma | FC010 | |
| Immersion Oil | Cargille | 16241 | |
| L-Glutamine Solution (200 mM) | Wisent Bioproducts | 609-065-EL | |
| MEM Non-Essential Amino Acids Solution (100X) | Gibco | 11140050 | |
| Micro Cover Glasses, Square, No. 11/2 22 x 22 mm | VWR | CA48366-227-1 | |
| Microscope Body, Eclipse Ti2-E | Nikon | ||
| Objective, CFI Plan Apo Lambda 60X Oil | Nikon | MRD01605 | |
| Penicillin-Streptomycin | Sigma | P4333 | |
| Spinning Disk, Crest Light V2 | CrestOptics | ||
| Spyder | Anaconda | ||
| Stage top incubator | Tokai Hit | ||
| Statistics Software, Prism | GraphPad | ||
| Tweezers, Style 2 | Electron Microscopy Sciences | 78326-42 |
References
- Mullins, R. D., Heuser, J. A., Pollard, T. D. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proceedings of the National Academy of Sciences. 95 (11), 6181-6186 (1998).
- Yang, C., Czech, L., Gerboth, S., Kojima, S., Scita, G., Svitkina, T. Novel Roles of Formin mDia2 in Lamellipodia and Filopodia Formation in Motile Cells. PLoS Biology. 5 (11), 317 (2007).
- Mogilner, A., Oster, G. Cell motility driven by actin polymerization. Biophysical Journal. 71 (6), 3030-3045 (1996).
- Mogilner, A., Oster, G. Force Generation by Actin Polymerization II: The Elastic Ratchet and Tethered Filaments. Biophysical Journal. 84 (3), 1591-1605 (2003).
- Pollard, T. D., Borisy, G. G. Cellular Motility Driven by Assembly and Disassembly of Actin Filaments. Cell. 112 (4), 453-465 (2003).
- Wu, C., et al. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 148 (5), 973-987 (2012).
- Steffen, A., et al. Rac function is crucial for cell migration but is not required for spreading and focal adhesion formation. Journal of cell science. 126, 4572-4588 (2013).
- Gupton, S. L., et al. Cell migration without a lamellipodium. The Journal of Cell Biology. 168 (4), 619-631 (2005).
- Dimchev, V., et al. Induced Arp2/3 Complex Depletion Increases FMNL2/3 Formin Expression and Filopodia Formation. Frontiers in Cell and Developmental Biology. 9, 634708 (2021).
- Leithner, A., et al. Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nature cell biology. 18 (11), 1253-1259 (2016).
- Giannone, G., Dubin-Thaler, B. J., Döbereiner, H. -. G., Kieffer, N., Bresnick, A. R., Sheetz, M. P. Periodic Lamellipodial Contractions Correlate with Rearward Actin Waves. Cell. 116 (3), 431-443 (2004).
- Dubin-Thaler, B. J., et al. Quantification of Cell Edge Velocities and Traction Forces Reveals Distinct Motility Modules during Cell Spreading. PLoS ONE. 3 (11), 3735 (2008).
- Suraneni, P., Rubinstein, B., Unruh, J. R., Durnin, M., Hanein, D., Li, R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. The Journal of cell biology. 197 (2), 239-251 (2012).
- Wang, C., et al. Deconvolution of subcellular protrusion heterogeneity and the underlying actin regulator dynamics from live cell imaging. Nature Communications. 9 (1), 1688 (2018).
- Dimchev, G., et al. Lamellipodin tunes cell migration by stabilizing protrusions and promoting adhesion formation. Journal of cell science. 133 (7), 239020 (2020).
- Burnette, D. T., et al. A role for actin arcs in the leading-edge advance of migrating cells. Nature cell biology. 13 (4), 371-381 (2011).
- Yamada, K. M., Kennedy, D. W. Dualistic nature of adhesive protein function: fibronectin and its biologically active peptide fragments can autoinhibit fibronectin function. The Journal of Cell Biology. 99 (1), 29-36 (1984).
- Cai, Y., et al. Nonmuscle Myosin IIA-Dependent Force Inhibits Cell Spreading and Drives F-Actin Flow. Biophysical Journal. 91 (10), 3907-3920 (2006).
- Humphries, M. J. Cell adhesion assays. Molecular Biotechnology. 18 (1), 57-61 (2001).
- Cavalcanti-Adam, E. A., Volberg, T., Micoulet, A., Kessler, H., Geiger, B., Spatz, J. P. Cell Spreading and Focal Adhesion Dynamics Are Regulated by Spacing of Integrin Ligands. Biophysical Journal. 92 (8), 2964-2974 (2007).
- Dubin-Thaler, B. J., Giannone, G., Döbereiner, H. -. G., Sheetz, M. P. Nanometer Analysis of Cell Spreading on Matrix-Coated Surfaces Reveals Two Distinct Cell States and STEPs. Biophysical Journal. 86 (3), 1794-1806 (2004).
- Gauthier, N. C., Fardin, M. A., Roca-Cusachs, P., Sheetz, M. P. Temporary increase in plasma membrane tension coordinates the activation of exocytosis and contraction during cell spreading. Proceedings of the National Academy of Sciences. 108 (35), 14467-14472 (2011).
- Wolfenson, H., Iskratsch, T., Sheetz, M. P. Early Events in Cell Spreading as a Model for Quantitative Analysis of Biomechanical Events. Biophysical Journal. 107 (11), 2508-2514 (2014).
- Guan, J. -. L., Berrier, A. L., LaFlamme, S. E. Cell Migration, Developmental Methods and Protocols. Methods in molecular biology. 294, 55-68 (2004).
- Raucher, D., et al. Phosphatidylinositol 4,5-Bisphosphate Functions as a Second Messenger that Regulates Cytoskeleton-Plasma Membrane Adhesion. Cell. 100 (2), 221-228 (2000).
- Machacek, M., Danuser, G. Morphodynamic Profiling of Protrusion Phenotypes. Biophysical Journal. 90 (4), 1439-1452 (2006).
- Zack, G. W., Rogers, W. E., Latt, S. A. Automatic measurement of sister chromatid exchange frequency. The journal of histochemistry and cytochemistry official journal of the Histochemistry Society. 25 (7), 741-753 (1977).
- Bardsley, W. G., Aplin, J. D. Kinetic analysis of cell spreading. I. Theory and modelling of curves. Journal of cell science. 61, 365-373 (1983).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved