A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Synthesis and Characterization of mRNA-Loaded Poly(Beta Aminoesters) Nanoparticles for Vaccination Purposes
In This Article
Summary
Here, a simple protocol is presented for producing mRNA nanoparticles based on poly(beta aminoester) polymers, easy to be tailored by changing the encapsulated mRNA. The workflow for synthesizing the polymers, the nanoparticles, and their in vitro essential characterization are also described. A proof-of-concept regarding immunization is also added.
Abstract
Vaccination has been one of the major successes of modern society and is indispensable in controlling and preventing disease. Traditional vaccines were composed of entire or fractions of the infectious agent. However, challenges remain, and new vaccine technologies are mandatory. In this context, the use of mRNA for immunizing purposes has shown an enhanced performance, as demonstrated by the speedy approval of two mRNA vaccines preventing SARS-CoV-2 infection. Beyond success in preventing viral infections, mRNA vaccines can also be used for therapeutic cancer applications.
Nevertheless, the instability of mRNA and its fast clearance from the body due to the presence of nucleases makes its naked delivery not possible. In this context, nanomedicines, and specifically polymeric nanoparticles, are critical mRNA delivery systems. Thus, the aim of this article is to describe the protocol for the formulation and test of an mRNA vaccine candidate based on the proprietary polymeric nanoparticles. The synthesis and chemical characterization of the poly(beta aminoesters) polymers used, their complexation with mRNA to form nanoparticles, and their lyophilization methodology will be discussed here. This is a crucial step for decreasing storage and distribution costs. Finally, the required tests to demonstrate their capacity to in vitro transfect and mature model dendritic cells will be indicated. This protocol will benefit the scientific community working on vaccination because of its high versatility that enables these vaccines to prevent or cure a wide variety of diseases.
Introduction
Infectious diseases have represented a severe threat to millions of human beings around the globe and are still one of the leading causes of death in some developing countries. Prophylactic vaccination has been one of the most effective interventions of modern society to prevent and control infectious diseases1,2. These critical milestones of science in 20th-century relevance have been remarked by the recent worldwide Covid-19 pandemic caused by the SARS-CoV-2 virus3. Recognizing the importance of having efficient vaccines to curtail the dissemination of the disease, cooperative efforts from all biomedical communities have successfully resulted in many prophylactic vaccines in the market in less than a year4.
Traditionally, vaccines were composed of attenuate (live, reduced virulence) or inactivated (death particles) viruses. However, for some diseases with no margin for safety errors, viral particles are not possible, and protein subunits are used instead. Nevertheless, subunits usually do not enable the combination of more than one epitope/antigen, and adjuvants are required to enhance vaccination potency5,6. Therefore, the need for novel vaccine types stands clear.
As demonstrated during the current pandemic, novel vaccine candidates based on nucleic acids can be advantageous in terms of avoiding long development processes and providing high versatility while producing, at the same time, a vital patient immunization. This is the case of mRNA vaccines, which were initially designed as experimental cancer vaccines. Thanks to their natural capacity to produce antigen-specific T-cell responses3,5,6,7. Being mRNA the molecule that encodes the antigenic protein, only changing the same, the vaccine can be rapidly tailored to immunize other variants of the same microorganism, different strains, other infectious microorganisms, or even become a cancer immunotherapeutic treatment. In addition, they are advantageous in terms of large-scale production costs. However, mRNA has a significant hurdle that hampers their naked administration: its stability and integrity are compromised in physiological media, full of nucleases. For this reason, the use of a nanometric carrier that protects it and vectorizes mRNA to the antigen-presenting cells is required2,8.
In this context, poly(beta aminoesters) (pBAE) are a class of biocompatible and biodegradable polymers that demonstrated a remarkable ability to complex mRNA in nanometric particles, thanks to their cationic charges9,10,11. These polymers are composed of ester bonds, which makes their degradation easy by esterases in physiological conditions. Among the pBAE library candidates, those functionalized with end cationic oligopeptides showed a higher capacity to form small nanoparticles to efficiently penetrate cells through endocytosis and transfect the encapsulated gene material. Furthermore, thanks to their buffering capacity, the acidification of the endosome compartment allows endosomal escape12,13. Namely, a specific kind of pBAE, including hydrophobic moieties on their backbone (the so-named C6 pBAE) to enhance their stability and end-oligopeptide combination (60% of polymer modified with a tri-lysine and 40% of the polymer with a tri-histidine) that selectively transfects antigen-presenting cells after parenteral administration and produce the mRNA encoded antigen presentation followed by mice immunization has been recently published14. In addition, it has also been demonstrated that these formulations could circumvent one of the main bottleneck steps of nanomedicine formulations: the possibility to freeze-dry them without losing their functionality, which enables long-term stability in soft dry environments15.
In this context, the objective of the current protocol is to make the procedure for the formation of the mRNA nanoparticles available to the scientific community by giving a description of the critical steps in the protocol and enabling the production of efficient vaccines for infectious diseases prevention and tumor treatment applications.
The following protocol describes the complete workout to synthesize oligopeptide end-modified poly(beta aminoesters) - OM-pBAE polymers that will further be used for nanoparticle synthesis. In the protocol, nanoparticles formulation is also included. In addition, critical steps for the success of the procedure and representative results are also provided to ensure that the resulting formulations accomplish the required quality control characterization features to define a positive or negative result. This protocol is summarized in Figure 1.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Synthesis of pBAE polymer with end oligopeptides (OM-pBAE)
- Polymerization of C6-pBAE
- Add 5-amino-1-pentanol (38 mmol; MW = 103.16 Da) 1-hexylamine (38 mmol; MW = 101.19 Da) into a round-bottom glass flask (100 mL). Then, add 1,4-butanediol diacrylate (82 mmol; MW = 198.22 Da).
- Pre-heat the silicone oil bath at 90 °C, place the round-bottom flask into the oil bath and stir the mixture with the aid of a magnetic stir bar overnight (~18 h). Then, take the product from the round-bottom flask and place it in the freezer at -20 °C.
NOTE: The product is in the form of a sticky powder and is taken out from the flask with the help of a spatula. It is essential to validate the structure of the obtained polymer through 1H-NMR. NMR spectra were recorded in a 400 MHz instrument (see Table of Materials) using chloroform-d and D2O as solvents. Around 10 mg of each poly(β-amino ester) were taken and dissolved in 1 mL of the deuterated solvent.
- Reaction with peptides to obtain OM-pBAE
- Add 25 mL of 0.1 M HCl to selected peptides, e.g., peptide Cys-His-His-His with Trifluoroacetic acid (TFA, 200 mg), into a previously weighed 50 mL centrifuge tube that can stand freeze-drying and manually stir overnight to obtain a clear solution.
- Freeze the solution at -80 °C for 1 h and freeze-dry the resulting peptide hydrochloride.
NOTE: Check whether the final weight corresponds to the theoretical value. - Make a solution of C6-pBAE (0.031 mmol) in dimethyl sulfoxide. Also, make a solution of the peptide hydrochloride (0.078 mmol) in dimethyl sulfoxide.
- Mix the two solutions in a screw cap tube and screw on the cap. Stir the mixture solution in a water bath with a controlled temperature of 25 °C for 20 h with a magnetic stir bar.
- Add the mixture to 7:3 (v/v) diethyl ether/acetone. Centrifuge the resulting suspension at 25,000 x g at 4 °C to remove the solvent. Next, wash the solid with 7:3 (v/v) diethyl ether/acetone twice. Then, dry the product under a vacuum (<0.2 atm).
- Make a solution of 100 mg/mL of the product in dimethyl sulfoxide. The resulting product is named C6-peptide-pBAE. It is essential to validate the structure of the obtained polymer through 1H-NMR to confirm the disappearance of the olefin signals associated with terminal acrylates. If not used, the polymer can be frozen at -20 °C.
2. Polyplexes formation
NOTE: All the procedures should be performed inside a conditioned room to maintain a constant temperature.
- Thaw the polymers C6-peptide-pBAE and vortex the solution.
- Pipette the polymer mix up and down and prepare a solution of 12.5 mM (V1) in sodium acetate (NaAc). Then, vortex the mixture and wait for 10 min.
- Prepare the mRNA at 0.5 mg/mL and mix by pipetting (V2).
NOTE: It is crucial to avoid vortexing the mRNA. - Vortex the polymer mixture at the final concentration to achieve a homogeneous solution between the polymer stock in DMSO and the acetate buffer.
NOTE: The final polymer concentration depends on the N/P (nitrogen to phosphate groups) ratio selected. The N/P ratio depends on each specific mRNA to be used. For eGFP encapsulation, for example, a 25:1 ratio was used, as previously reported14. - Mix the genetic material solution and C6-peptidepBAE solution (25x of the mRNA concentration) in a ratio of 1:1 (Vi = V1 + V2).
NOTE: C6-peptide-pBAE is loaded in a microcentrifuge tube where the mRNA is added by pipetting up and down for mixing. Once prepared, the polyplex, the nucleic acid, and C6-peptide-pBAE concentrations are half diluted. - Incubate at 25 °C for 30 min in a thermoblock. Precipitate with 1:2 RNase free water by adding the sample to a pre-loaded microcentrifuge tube with water.
- Include the excipients. Add the same volume as the mixture of mRNA and pBAE (Vi) in HEPES 20 mM and sucrose 4% by pipetting up and down. At this point, the sample has been diluted 3x.
3. Polyplexes lyophilization
- Instantaneously, freeze at -80 °C freezer of the previous polyplex solution for 1 h.
- Perform the primary drying by following the steps: (1) 1 h at -60 °C and 0.001 hPa; (2) 1 h at -40 °C and 0.0001 hPa; (3) 4 h at -20 °C and 0.0001 hPa; (4) 12 h at 5 °C and 0.0001 hPa.
- Store at -20 °C immediately to avoid rehydration until use.
4. Polyplex resuspension
NOTE: This protocol describes the process used to reconstruct the lyophilized C6-peptide-pBAE nanoparticles for their further use either for characterization, in vitro, or in vivo analysis.
- Take the lyophilized nanoparticles from -20 °C just at the moment of use and rapidly add the corresponding amount of depyrogenated (DEPC) water to redisperse the solid to achieve the desired concentration.
NOTE: The volume will be the same as the initial volume of the nanoparticles if the same concentration is envisaged, but it will be indicated for each experiment. - Pipette gently until total resuspension, accompanying the liquid with the pipette.
NOTE: Assure that no material remains on the wall of the vial. - Once dissolved, pipette up and down vigorously, avoiding bubbles.
NOTE: The sample should have a transparent to translucent aspect, which is more evident at higher concentrations - Store the samples on ice or at 4 °C for a maximum period of 24 h once reconstituted. Avoid freezing.
5. Polyplex characterization
- Dynamic light scattering
NOTE: Hydrodynamic diameter (nm), polydispersity index (PDI), and surface charge of nanoparticles were measured at 25 °C, 633 nm laser wavelength, and 173° signal detector, using a Zeta potential analyzer (see Table of Materials).- Hydrodynamic diameter (nm)
- Prepare the nanoparticles mixing mRNA and pBAE thoroughly by pipetting the gene material to the polymer fraction to a final concentration of mRNA of 0.25 mg/mL as previously described above (step 2).
- Pre-rinse a microcuvette with filtered dilution media to completely clean it from impurities. Then, fill the cuvette with at least 50 µL of the sample solution and cap it. Next, introduce the sample inside the DLS instrument, ensure that the cell is correctly inserted.
NOTE: The dilution media is filtered using a 0.22 µm syringe filter. - Open the Standard Operation Procedure (SOP) file created and introduce the desired Sample Name. Select Size Measurement.
NOTE: All the parameters to create an SOP for size measurements are provided in Table 1. - Run the particle size measurement using DLS by clicking on Play.
NOTE: After the triple beep sounds, the analysis is complete. - Select the results corresponding to the sample on the measurement sheet to obtain the average particle size, average PDI, standard deviation, and graphics. Once done, remove the cell from DLS equipment.
- Remove the sample, keep it for zeta-potential analysis, and clean and rinse the cell with deionized water. Finally, dry the cleaned cuvette under a compressed air gas stream.
- Surface charge (mV)
- Prepare the nanoparticles mixing mRNA and pBAE thoroughly by pipetting the gene material to the polymer fraction to a final concentration of mRNA of 0.25 mg/mL as previously described above (step 2, Polyplex formation). Next, freeze and dry the solution.
NOTE: In this case, for the measurement of surface charge, the freeze-drying of the sample is strongly recommended before performing the measures to redisperse the sample in the appropriate buffer, simulating body conditions of pH and electrolyte concentration. - Resuspend the lyophilized sample using water. Dilute the sample 1/10 in water (final concentration 0.025 mg/mL).
- Pre-rinse a disposable folded capillary cell (zeta potential cuvette) with filtered dilution media to completely clean it from impurities. Then, fill the cuvette with diluted nanoparticles, using a 1 mL syringe, and cap both sides of the fillers.
NOTE: The nanoparticle dispersion has to fill the volume available in the cuvette (~1 mL), taking special attention to the bubble's formation, which could perturb measures. - Introduce the sample inside DLS; ensure that the cell is correctly inserted.
- Open the SOP file created for zeta potential analysis and introduce the desired Sample Name. Select Zeta-Potential Measurement.
NOTE: Parameters to create an SOP for zeta potential measurements are given in Table 2. - Run zeta-potential measurement using DLS by clicking on Play.
NOTE: After the triple beep sound, the analysis is complete. - Select the results corresponding to the sample on the measurement sheet to obtain the average zeta-potential, standard deviation, and graphics. Once done, remove the cell from the DLS equipment.
- Remove the sample and keep it if it is necessary. Next, clean and rinse the cuvette cell with deionized water, followed by ethanol and water again. Finally, dry the cleaned cuvette under a compressed air gas stream.
NOTE: To analyze the data, use the recommended software.
- Prepare the nanoparticles mixing mRNA and pBAE thoroughly by pipetting the gene material to the polymer fraction to a final concentration of mRNA of 0.25 mg/mL as previously described above (step 2, Polyplex formation). Next, freeze and dry the solution.
- Hydrodynamic diameter (nm)
- Nanoparticle Tracking Analysis (NTA)
NOTE: Hydrodynamic diameter (nm) and concentration of nanoparticles were measured at 25 °C, 488 nm laser wavelength using a Nanoparticle Tracking Analyzer (see Table of Materials).- NTA and sample preparation
- Switch on the computer and the equipment. First, check whether all the components are plugged in correctly. Next, open the recommended software. The software will check whether all the accessories are correctly connected.
- Connect the Top plate, here, the O-ring cell, into the laser module.
NOTE: Do not overscrew this part. - First load the chamber with buffer and/or deionized water with a 1 mL syringe. Repeat the procedure at least two times. Avoid introducing bubbles in the chamber.
- Prepare the sample by diluting 1/1000 in deionized water from the concentration used in DLS size measurement. Prepare at least 1 mL and load it into a 1 mL syringe, avoiding introducing bubbles.
- Load the samples into the chamber using a syringe. Then, plug the laser module into the NTA big chamber.
NOTE: Big chamber denotes the space where the chamber is placed for the measurement.
- Image optimization
- First, in the Hardware, check whether the Camera and the proper laser are selected.
NOTE: Here, there is only one camera and the Blue Laser 488 nm. - Start with the camera level at 0 and press Start Camera on the Capture window.
NOTE: At this point, adjust the following parameters. Beam position (it can be moved up and down on the screen). Camera Level: avoid oversaturated pixels. Focus (lateral wheel): try to focus as better as possible. It is better to have particles with a halo instead of unfocused particles. Concentration: Adjust the concentration to have between 10 to 100 particles per field.
- First, in the Hardware, check whether the Camera and the proper laser are selected.
- Video recording
- In the software, go to the Measurement Selection- SOP window (down left) and select Standard Measurement.
NOTE: Program to perform three measures of 30 s each (to enable the software to perform mean and standard deviation calculations). Provide a name and a folder path (on Base filename) to save the records of the sample. When the SOP parameters are correctly set up, press Create and Run Script. Once the first replicate is measured, the program will ask to add a new sample. Then, another fraction of the sample must be introduced in the chamber by pushing the syringe's plunger. Finally, repeat a third time to analyze the third replicate.
- In the software, go to the Measurement Selection- SOP window (down left) and select Standard Measurement.
- Video processing
- Readjust the Screen Gain and Detection Threshold once the three measures are finished to analyze the measured particles. At this moment, around 100 particles should appear in each frame to perform the analysis (high red crosses and low blue crosses are expected).
- Export the results in a pdf file after the analysis is completed. In addition, it is possible to export as videos and excel files.
- Cleaning the NTA
NOTE: Close all the opened measurements before studying the following sample; since the generated files are enormous and depending on the computer, it isn't easy to maintain more than one open.- Clean the NTA chamber by repeatedly flushing water before performing the subsequent measurement until no particles are observed; subsequently, flush the buffer used (PBS) to continue with the measures.
- Flush air inside the chamber and dry it with microscopic grade paper once the last measurement of the day is completed.
- Characterize the size and shape by Transmission Electronic Microscopy (TEM). Prepare the samples. 30 µL of the final volume is enough for TEM characterization.
NOTE: Both fresh and lyophilized - after resuspension- nanoparticles can be measured. - Drop 10 µL of sample on a carbon-coated copper grid. Let it dry for 10 min. Remove the excess liquid, if necessary, by softly tapping on filter paper.
- Drop 10 µL of uranyl acetate (2% w/v) solution for negative staining. Let it dry for 1 min. Remove the liquid excess, if necessary, by softly tapping on filter paper.
- Introduce the sample in the microscope and scan (voltage operation 80 kV).
NOTE: Appropriate software can be used for further analysis of the images.
- NTA and sample preparation
- Encapsulation efficiency
- Sample preparation
- Prepare the nanoparticles at the desired concentration and freeze and dry them. Then, resuspend the nanoparticles and dilute them at a final concentration of 6 µg/mL.
- Prepare 1x TE buffer from the 20x stock solution.
NOTE: Vt (Total volume) = (number of samples x 100 µL x 4) (number of samples x 290 µL) + 2 mL. - Prepare TE buffer with 3 µg/µL of Heparin from the stock of 100 µg/µL.
NOTE: Vt = Number of samples x 50 µL x 2. - Prepare the standards
- Prepare an RNA standard ranging from 0.2 µg/mL to 0.025 µg/mL in 1x Tris-EDTA (TE) buffer (Table S1A).
NOTE: Use mRNA in the RNA standard in case that differs from the Ribosomal RNA. - Prepare RNA:pBAE standard with Heparin (Table S1B). Prepare Heparin standard (Table S1C).
- Prepare a 96-well plate as follows.
- Load 295 µL of TE buffer in the first lane of a 96-well plate (as many wells as samples to analyze). Then, load 50 µL of 1x TE buffer into lanes B and C (duplicates for each sample).
- Load 50 µL of 1x TBE buffer with 3 µg/µL of Heparin into lanes D and E (duplicates for each sample). Next, load 100 µL of each standard in lanes F, G, and H (duplicates for each standard).
- Load 5 µL of each sample in lane A (concentration in each well is 0.1 µg/mL). Mix properly by pipetting and load 50 µL of each sample on the four wells below (two of them containing 50 µL of buffer 1x TE and two more containing 1x TE buffer with 3 µg/µL of Heparin).
- Incubate the sample for 30 min at 37 °C. Prepare RiboGreen reagent as per the manufacturer's protocol (see Table of Materials) in 1x TE buffer.
NOTE: Dilute 1:200 and vortex. Protect from light. Vt = Number of full wells x 100 µL. - Load 100 µL of RiboGreen solution into each well. Detect fluorescence using the microplate reader with an excitation wavelength of 500 nm and an emission wavelength of 525 nm.
- Results analysis
- Prepare the three calibration curves
NOTE: First, Ribosomal RNA standard. Interpolate in this calibration curve the samples that do not contain Heparin. Second, mRNA:pBAE with Heparin. Interpolate in this calibration curve the sample that has Heparin. Third, Heparin. Used to assure the working concentration is in the linear range. - Calculate encapsulation efficiency (EE%) as follows:
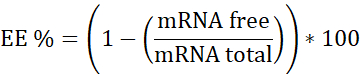
- Prepare the three calibration curves
- Sample preparation
6. In vitro characterization
- Fluorescence microscopy for qualitative assessment
- Place the 96-well plate onto the microscope after 24 h of transfection. Start visualizing cells with the 10x objective.
NOTE: HeLa cells are used in this case. Cells are localized using the transmission mode (bright field), and the focus must be adjusted at this time.- First, apply a white balance to reference the software about the background of the samples. Then, acquire an image to overlay it with the fluorescence image during the analysis.
- Change the microscope mode to reflection mode (fluorescence) and move the filter wheel to the blue laser (488 nm) to visualize eGFP.
NOTE: At this point, the Exposure time and Gain must be adjusted. Gain must be adjusted in values between 3 and 5 to avoid artifacts and excess on background signal. The adjustment of the exposure time depends on the transfection efficiency (from ms to 1 s). - Acquire images for all the conditions or wells employing the same exposure time to compare all the samples.
- Place the 96-well plate onto the microscope after 24 h of transfection. Start visualizing cells with the 10x objective.
- Flow cytometry (FC) for quantitative assessment
- Use a multichannel pipette to prepare the 96-well plate for the flow cytometry experiment.
NOTE: When working with semi-adherent cells, such as in the JAWSII cell line, recover all the media volume and store it into another 96-well plate. Do not aspire to this media since it might contain a significant number of transfected cells. - Clean the cells (HeLa cells are used here) with 100 µL/well of 1x PBS and aspire it. Next, add 25 µL/well of trypsin and incubate it at 37 °C for 5 min.
- Once the cells are detached, add previously recovered media to stop the trypsin action and fix the cells, adding 31.25 µL/well of formalin 10% for 20 min (final concentration 2.5%).
NOTE: At this point, it is crucial to visualize the state of the cells by microscopy to ensure the detachment of the cells. It is highly recommended to pipette up and down several times to help the cells detach and avoid their agglomeration. This will produce a reliable result. - Turn on the flow cytometer and the software. In the software, set up the proper conditions for the experiment (type of plate, sample volume, and other parameters such as shaking, rinsing between samples.
NOTE: For step 6.2.5, the flow rate should be 120 µL/min. Mix one cycle every 1 min and rinse one cycle every one well. Mixing is essential to maintain the cells dispersed in the media and permit a better analysis of individual cells. In addition, rinsing is necessary to clean the microfluidics of the cytometer between samples. Finally, check whether there are enough buffers and whether all components are correctly connected. - Set up the appropriate parameters to quantify the percentage of positively transfected cells.
- First, view the flow data on (Forward scattered light) FSC versus SCC (Side scattered light) scatter plot to distinguish the cells from the debris. Then, another scatter plot comparing the amplitude (FSC-A) versus height (FSC-H) is plotted to gate and discriminate individual cells.
NOTE: The untreated population allows to gate the populations corresponding to positively transfected cells. Then, quantification of the positively transfected cells is performed on the proper channel on a histogram plot. Therefore, the positively transfected cells must be represented as a continuum of events on the histogram.
- First, view the flow data on (Forward scattered light) FSC versus SCC (Side scattered light) scatter plot to distinguish the cells from the debris. Then, another scatter plot comparing the amplitude (FSC-A) versus height (FSC-H) is plotted to gate and discriminate individual cells.
- Use a multichannel pipette to prepare the 96-well plate for the flow cytometry experiment.
7. In vitro functionality tests: capacity to activate model immune cells by using ovalbumin (OVA) as antigenic model mRNA
- Plate 10,000 cells/well (JAWSII cells are used here) in a 96-well plate the day before transfection.
NOTE: Plate as many wells as required to have triplicates for each condition. Let the cells attach to the plate at least for 12 h (an overnight incubation is recommended). - Prepare pBAEs NPs as described in the NOTE below to transfect 0.6 µg mRNA per well.
NOTE: As a positive control, transfect the cells using a transfection reagent. Here, we used 0.1 µg mRNA per well and 0.25 µL of the transfection reagent per well. mRNA codifying for OVA was purchased (see Table of Materials). It is polyadenylated, modified with 5-methoxyuridine, and optimized for mammalian systems, and is protected through an end-capping with Cap1 structure, a proprietary method from the supplier. - Incubate the 96-well plate for 24 h in a dry air incubator at 37 °C and 5% CO2.
NOTE: After this time, do not aspire the media since it might contain a certain number of cells. Recover the cells and save them in another well or plate. - Wash the cells remaining on the well with 25 µL of 1x PBS. Next, aspire it and add 25 µL of trypsin and incubate the plate for 5 min at 37 °C to detach the cells. Stop the trypsin reaction by adding the previously recovered media onto the correspondent well.
- Centrifuge the plate at 400 x g for 5 min at 4 °C. Aspire the media. Add 50 µL/well of a 1x PBS and 2.5% of formalin. Incubate it at 4 °C for 20 min to fix cells.
- Repeat steps 7.5 and 7.5.1. Then, add 50 µL/well of 1x PBS and 3% BSA (Blocking buffer) and incubate for 30 min at 4 °C. Again, repeat steps 7.5 and 7.5.1, and then add 50 µL/well of the primary antibody (mouse α-OVA) in 1x PBS and 3% BSA and incubate for 30 min at 4 °C.
- Repeat steps 7.5 and 7.5.1, and then wash the cells with 50 µL/well of 1x PBS. Aspire media, and then add secondary antibodies solution (α-mouse-AlexaFluor488/PerCP and Cy5.5-CD11b/APC-CD86) in 1x PBS and 3% BSA. Incubate it for 1 h at 4 °C.
- Centrifuge the plate at 400 x g for 5 min. Then, wash the cells with 50 µL/well of 1x PBS. Next, centrifuge (400 x g for 5 min), aspire, and then resuspend the cells in 100 µL/well of 1x PBS and 2.5% Formalin.
- Analyze it by flow cytometry, as described above (step 6.2).
Access restricted. Please log in or start a trial to view this content.
Results
Polymer synthesis and characterization
The OM-pBAE synthesis procedure is given in Figure 2. As Figure 2A shows, the first step to obtain the OM-pBAE is to synthesize the C6-pBAE by adding the amines (1-hexylamine and 5-amino-1-pentanol, ratio 1:1) to the diacrylate (1,4-butanediol diacrylate). This reaction is carried out at 90 °C for 20 h and with constant stirring. Afterward, a solution of oligopeptides is added to a solution ...
Access restricted. Please log in or start a trial to view this content.
Discussion
After the outbreak of the Covid-19 pandemic last year, the importance of vaccines in terms of infectious disease control has manifested as a critical component8. Efforts from scientists worldwide have enabled the release to the market of many vaccines. For the first time in history, mRNA vaccines have demonstrated their previously hypothesized success, thanks to their rapid design because of their capacity to adapt to any novel antigen within some months5,
Access restricted. Please log in or start a trial to view this content.
Disclosures
Authors have nothing to disclose nor any conflicts of interest.
Acknowledgements
Financial support from MINECO/FEDER (grants SAF2015-64927-C2-2-R, RTI2018-094734-B-C22, and COV20/01100) is acknowledged. CGF acknowledged her IQS PhD Fellowship.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 1,4-butanediol diacrylate | Sigma Aldrich | 123048 | |
| 1-hexylamine | Sigma Aldrich | 219703 | |
| 5-amino-1-pentanol | Sigma Aldrich | 411744 | |
| Acetone | Panreac | 141007 | |
| CD11b antibody | BD | 550993 | |
| CD86 antibody | Bioligend | 105007 | |
| Chlor hydroxhyde | Panreac | 181023 | |
| Chloroform-d | Sigma Aldrich | 151823 | |
| Cys-His-His-His peptide | Ontores | Custom | |
| Cys-Lys-Lys-Lys peptide | Ontores | Custom | |
| D2O | Sigma Aldrich | 151882 | |
| DEPC reagent for Rnase free water | Sigma Aldrich | D5758 | This reagent is important to treat MilliQ water to remove any RNases of the buffers |
| Diethyl eter | Panreac | 212770 | |
| dimethyl sulfoxide | Sigma Aldrich | 276855 | |
| HEPES | Sigma Aldrich | H3375 | |
| mRNA EGFP | TriLink Technologies | L-7601 | |
| mRNA OVA | TriLink Technologies | L-7610 | |
| RiboGreen kit | ThermoFisher | R11490 | |
| sodium acetate | Sigma Aldrich | 71196 | |
| sucrose | Sigma Aldrich | S0389 | |
| Trifluoroacetic acid | Sigma Aldrich | 302031 | |
| Trypsin-EDTA | Fisher Scientific | 11570626 | |
| α-mouse AlexaFluor488 antibody | Abcam | Ab450105 | |
| Equipment | |||
| Nanoparticle Tracking Analyzer | Malvern Panalytical | NanoSight NS300 | |
| Nuclear Magnetic Ressonance Spectrometer | Varian | 400 MHz | |
| ZetaSizer | Malvern Panalytical | Nano ZS | For zeta potential and hydrodynamic size determination |
| Software | |||
| NanoSight NTA software | Malvern Panalytical | MAN0515-02-EN-00 | |
| NovoExpress Software | Agilent | Not specified | |
| ZetaSizer software | Malvern Panalytical | DTS Application | To analyze surface charge and hydrodynamic sizes |
References
- Chumakov, K., Benn, C. S., Aaby, P., Kottilil, S., Gallo, R. Can existing live vaccines prevent COVID-19. Science. 368 (6496), 1187-1188 (2020).
- Zhang, C., Maruggi, G., Shan, H., Li, J. Advances in mRNA vaccines for infectious diseases. Frontiers in Immunology. 10, 1-13 (2019).
- Wherry, E. J., Jaffee, E. M., Warren, N., D'Souza, G., Ribas, A. How did we get a COVID-19 vaccine in less than 1 year. Clinical Cancer Research. 27 (8), 2136-2138 (2021).
- Folegatti, P. M., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. The Lancet. 396 (10249), 467-478 (2020).
- Geall, A. J., Mandl, C. W., Ulmer, J. B. RNA: The new revolution in nucleic acid vaccines. Seminars in Immunology. 25 (2), 152-159 (2013).
- Ulmer, J. B., Geall, A. J. Recent innovations in mRNA vaccines. Current Opinion in Immunology. 41, 18-22 (2016).
- Kranz, L. M., et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 534 (7607), 396-401 (2016).
- Milane, L., Amiji, M. Clinical approval of nanotechnology-based SARS-CoV-2 mRNA vaccines: impact on translational nanomedicine. Drug Delivery and Translational Research. 1 (4), 3(2020).
- Green, J. J., Langer, R., Anderson, D. G. A combinatorial polymer library approach yields insight into nonviral gene delivery. Accounts of Chemical Research. 41 (6), 749-759 (2008).
- Guerrero-Cázares, H., et al. Biodegradable polymeric nanoparticles show high efficacy and specificity at DNA delivery to human glioblastoma in vitro and in vivo. ACS Nano. 8 (5), 5141-5153 (2014).
- Kozielski, K. L., Tzeng, S. Y., Hurtado De Mendoza, B. A., Green, J. J. Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells. ACS Nano. 8 (4), 3232-3241 (2014).
- Segovia, N., Dosta, P., Cascante, A., Ramos, V., Borrós, S. Oligopeptide-terminated poly(β-amino ester)s for highly efficient gene delivery and intracellular localization. Acta Biomaterialia. 10 (5), 2147-2158 (2014).
- Dosta, P., Segovia, N., Cascante, A., Ramos, V., Borrós, S. Surface charge tunability as a powerful strategy to control electrostatic interaction for high efficiency silencing, using tailored oligopeptide- modified poly (beta-amino ester)s (PBAEs). Acta Biomaterialia. 20, 82-93 (2015).
- Fornaguera, C., et al. mRNA delivery system for targeting antigen-presenting cells in vivo. Advanced Healthcare Materials. 7 (17), 180033(2018).
- Fornaguera, C., Castells-Sala, C., Lázaro, M. Á, Cascante, A., Borrós, S. Development of an optimized freeze-drying protocol for OM-PBAE nucleic acid polyplexes. International Journal Pharmaceutics. 569, (2019).
- Fornaguera, C., Solans, C. Analytical methods to characterize and purify polymeric nanoparticles. International Journal of Polymer Science. , (2018).
- Fornaguera, C., Solans, C. Characterization of polymeric nanoparticle dispersions for biomedical applications: size, surface charge and stability. Pharmaceutical Nanotechnology. 6 (3), 147-164 (2018).
- Sahin, U., Karikó, K., Türeci, Ö MRNA-based therapeutics-developing a new class of drugs. Nature Reviews Drug Discovery. 13 (10), 759-780 (2014).
- Fan, Y. N., et al. Cationic lipid-assisted nanoparticles for delivery of mRNA cancer vaccine. Biomaterials Science. 6 (11), 3009-3018 (2018).
- Le Moignic, A., et al. Preclinical evaluation of mRNA trimannosylated lipopolyplexes as therapeutic cancer vaccines targeting dendritic cells. Journal of Controlled Release. 278, 110-121 (2018).
- Banerji, A., et al. mRNA vaccines to prevent COVID-19 disease and reported allergic reactions: Current evidence and approach. Journal of Allergy and Clinical Immunology: In Practice. 9 (4), 1423-1437 (2021).
- Kaczmarek, J. C., Kowalski, P. S., Anderson, D. G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Medicine. 9 (1), 1-16 (2017).
- Dosta, P., et al. Delivery of anti-microRNA-712 to inflamed endothelial cells using poly(β-amino ester) nanoparticles conjugated with VCAM-1 targeting peptide. Advanced Healthcare Materials. , 1-11 (2021).
- Segovia, N., et al. Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer. Advanced Healthcare Materials. 4 (2), 271-280 (2015).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved