Method Article
Characterization of Neuromuscular Junctions in Mice by Combined Confocal and Super-Resolution Microscopy
* These authors contributed equally
In This Article
Summary
This protocol describes a method for morphometric analysis of neuromuscular junctions by combined confocal and STED microscopy that is used to quantify pathological changes in mouse models of SMA and ColQ-related CMS.
Abstract
Neuromuscular junctions (NMJs) are highly specialized synapses between lower motor neurons and skeletal muscle fibers that play an essential role in the transmission of molecules from the nervous system to voluntary muscles, leading to contraction. They are affected in many human diseases, including inherited neuromuscular disorders such as Duchenne muscular dystrophy (DMD), congenital myasthenic syndromes (CMS), spinal muscular atrophy (SMA), and amyotrophic lateral sclerosis (ALS). Therefore, monitoring the morphology of neuromuscular junctions and their alterations in disease mouse models represents a valuable tool for pathological studies and preclinical assessment of therapeutic approaches. Here, methods for labeling and analyzing the three-dimensional (3D) morphology of the pre- and postsynaptic parts of motor endplates from murine teased muscle fibers are described. The procedures to prepare samples and measure NMJ volume, area, tortuosity and axon terminal morphology/occupancy by confocal imaging, and the distance between postsynaptic junctional folds and acetylcholine receptor (AChR) stripe width by super-resolution stimulated emission depletion (STED) microscopy are detailed. Alterations in these NMJ parameters are illustrated in mutant mice affected by SMA and CMS.
Introduction
The neuromuscular junction (NMJ) is a complex structure composed of a motor axon terminal, a perisynaptic Schwann cell, and a skeletal myofiber portion involved in the transmission of chemical information and coupling of lower motor neuron activity to muscle contraction. In mammals, the morphology of the neuromuscular junction changes during development, adopting a typical pretzel-like shape after maturation, with differences in shape and complexity between species, and shows some degree of plasticity in response to physiological processes such as exercise or aging1,2,3,4. The postsynaptic motor endplate forms membrane invaginations named junctional folds, where the upper part containing acetylcholine receptors (AChR) is in close contact with the presynaptic terminal axon branch5.
Morphological and functional changes in neuromuscular junctions contribute to the pathophysiology of several neurodegenerative disorders such as spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS), myopathies like Duchenne muscular dystrophy (DMD), congenital myasthenic syndromes (CMS), myasthenia gravis (MG) and centronuclear myopathies (CNM), and aging-associated sarcopenia3,6,7,8,9,10,11,12. In these diseases, NMJ structural alterations such as endplate fragmentation, reduced postsynaptic junctional fold size and/or denervation are observed. The pathology of NMJs can be a primary or early event during disease progression or appear more lately as a secondary event contributing to the clinical manifestations. In any case, monitoring the morphology of NMJs in animal models of these diseases represents a valuable parameter to study pathological changes and assess the efficacy of potential treatments.
The morphology of neuromuscular junctions is usually analyzed by techniques using confocal microscopy2,13,14,15 or electron microscopy5,16, with their inherent limitations such as resolution or technical difficulties, respectively. More recently, super-resolution microscopy was also used to visualize particular regions of the NMJ, such as presynaptic active zones or AChR distribution on the postsynaptic membrane16,17,18, as an alternative or complementary approach to ultrastructural analysis by electron microscopy.
This protocol aims to provide a detailed and reproducible method to assess NMJ morphological parameters by combining fluorescence confocal and stimulated emission depletion (STED) microscopy. Important features of the presynaptic and postsynaptic endplates, such as volume, area, relative tortuosity, AChR stripe width, and axon terminal distribution in innervated teased muscle fibers of mouse gastrocnemius and tibialis anterior were quantified in the context of normal and diseased conditions. In particular, NMJ defects were exemplified in the Smn2B/- mouse model of spinal muscular atrophy, a neuromuscular disease with motor neuron degeneration caused by mutations in the SMN1 gene11,19, and in a collagen-like tail subunit of asymmetric acetylcholinesterase knockout (ColQDex2/Dex2 or ColQ-KO) mice, as a model of the congenital myasthenic syndrome20,21,22.
Protocol
Care and manipulation of mice were performed according to national and European legislation on animal experimentation and approved by the institutional ethical committee. Males and females of Smn2B/- (C57Bl/6J background) and ColQDex2/Dex2 (B6D2F1/J background) mice at 3- and 6-weeks of age, respectively, were used in the study.
1. Euthanasia of mice and dissection of muscles: tibialis anterior and gastrocnemius
- Proceed to mouse anesthesia by intraperitoneal injection of a ketamine (87.5 mg/kg)/xylazine (12.5 mg/kg) mixed solution (0.1 mL/20 g of body weight) prior to euthanasia by cervical dislocation.
NOTE: Since SMA and ColQ-CMS affect individuals independently of their sex, male and female mice were used in the present protocol. - Remove hindlimbs hair using a small electric shaver and rinse the legs with 70% ethanol.
NOTE: The dissection procedure will differ for each muscle. For dissection of tibialis anterior (TA), follow steps 1.2.1-1.2.3, and for gastrocnemius (GA) (medial and lateral parts), follow steps 1.2.4-1.2.6. Handle the muscles gently to prevent tissue damage and crushing or stretching them during dissection.- Place the mouse in the supine position.
- Make a skin incision of 5 mm with sharp-blunt scissors along the antero-external part of the distal hindlimb, parallel to the tibia, to expose the muscle. Use extra-thin scissors to remove the fascia.
- Cut the distal tendon first (close to the paw) and then the proximal tendon (close to the knee) using extra-thin scissors and a curved thin forceps. Handle the muscle carefully to avoid damage to myofibers and nerves.
NOTE: The proximal tendon must be sectioned as close as possible to the bone to harvest the entire muscle. - Place the mouse in the prone position, use sharp-blunt scissors to make a skin incision from the upper part of the distal hindlimb posterior compartment down to the paw, and remove the skin.
- Grasp the Achilles tendon with medium serrated forceps, cut it with an extra-thin scissor and gently separate the GA from the surrounding tissue back to its proximal insertion.
- On the proximal side, insert the medium serrated forceps into the pocket formed between the biceps femoris (BF) and the GA. Separate the two muscles to cut the GA tendon as close as possible to the bone insertion with an extra-thin scissor.
- For tissue fixation, place each muscle into a 2 mL microcentrifuge tube containing 1 mL of 4% w/v paraformaldehyde (PFA) solution diluted in phosphate buffer saline (PBS without Ca2+Mg2+) and keep at 4 °C for 18-24 h.
CAUTION: Paraformaldehyde and formaldehyde are toxic and must be handled in a chemical fume hood with adequate protective equipment. - Next day, wash the fixed muscles 3x for 5 min with PBS in 12-well plates by shaking gently at room temperature (RT) inside a chemical fume hood.
NOTE: The protocol can be stopped at this step and continued within a month. In this case, add PBS supplemented with 0.01% sodium azide to store samples at 4 °C. - Tease each muscle in small fiber bundles of about 1 mm wide using two fine serrated forceps.
NOTE: It is crucial to manipulate muscles very gently with the forceps, without excessive force, to prevent tissue damage during teasing.- Dissociate the TA muscle into 3 or 4 bundles depending on its size.
- For GA, separate the medial and lateral parts of the muscle and then dissociate each part into 4-5 bundles depending on their size.
2. Immunostaining
- Proceed with muscle fiber permeabilization: Transfer muscle bundles into 24-well plates containing 1% (v/v) Triton X-100 in PBS and keep them under gentle agitation (50 rpm) for 1 h at RT or 5 h at 4 °C.
NOTE: Split the muscle bundles between two plates to proceed with separate immunostainings and to minimize the risk of antibody confusion. Do not split them into more than two wells (1 well/plate); otherwise, the number (N) of NMJs that are representative of their general status in the analyzed muscle may be insufficient. - Wash the samples 3x for 5 min with PBS at RT and incubate them with a blocking solution composed of 4% bovine serum albumin (BSA) in PBS/Triton X-100 1% for 4 h at 4 °C, under gentle agitation (50 rpm).
NOTE: Do not use an aspiration pump during the washing steps, but rather aspirate the solution manually with a 200 µL pipette and small-sized tips (the reference is indicated in the Table of Materials). - Incubate the samples overnight (O/N) at 4 °C under gentle agitation (50 rpm) with the blocking solution indicated in step 2.2 containing primary monoclonal antibodies against either neurofilament M (NF-M, 2H3, dilution 1/200) or synaptic vesicle glycoprotein 2 (SV2, dilution 1/200) to label presynaptic axon terminals or active zones, respectively.
- Next day, wash the muscle bundles 3x for 5 min in PBS under agitation (50 rpm).
- For confocal imaging: Incubate the muscle bundles with secondary anti-mouse antibodies conjugated with a red-emitting fluorophore (F594) (dilution 1/500) and α-bungarotoxin conjugated with a green-emitting fluorophore (α-BTX-F488) (dilution 1/1000) in PBS for 2 h at RT under agitation (50 rpm).
- For STED imaging: Incubate the muscle bundles with secondary anti-mouse antibodies conjugated with a green-emitting fluorophore (F488) (dilution 1/500) and α-bungarotoxin conjugated with a far-red-emitting fluorophore characterized by high photostability (α-BTX-F633) (dilution 1/1000) in PBS for 2 h at RT under agitation (50 rpm).
NOTE: Do not expose the samples to light during incubation to avoid photobleaching.
- Wash the labeled muscle bundles 3x for 5 min with PBS under agitation (50 rpm) and place them on a slide with a mounting medium.
NOTE: Place a maximum of 4 to 5 muscle bundles per slide to allow sealing. - Add a grade #1.5 (or #1.5H) glass coverslip (0.17 mm thickness) on the top, and place cylindrical magnets on both sides of the slide to apply pressure and flatten the muscles.
- Keep the slides protected from light O/N at 4°C. Seal the slides permanently with nail polish.
3. Image acquisition
- Acquisitions by a confocal microscope
NOTE: Images were collected with an inverted laser-scanning confocal microscope using a 63x magnitude oil immersion objective (HCX Plan Apo CS, 1.4 numerical aperture (NA)).- For blinded analysis, let a person not involved in the analysis code each slide with a given number. Remain blinded to the experimental groups until the quantification of NMJ parameters is complete for all samples.
- Launch the microscope software in Configuration Mode > machine.xlhw (Supplemental Figure 1).
- Place the slide on the microscope stage and find the observation plane within the sample by looking under DAPI wide-field fluorescence illumination with the DAPI filter set.
- Click on Open Project > New Projects and create a folder to store image acquisitions (Supplemental Figure 1).
NOTE: Create a new project for each NMJ to limit folder size and prevent computer memory issues. - To manage acquisition parameters, click on the Acquisition tab window and set the confocal pinhole to 1.0 Airy unit and laser power to optimize the gain and offset levels for the green/F488 (α-BTX) fluorescence using a 488 nm laser at the endplate that has to be imaged (Live mode ON).
- Next, optimize the red/F594 (NF-M or SV2) fluorescence acquisition using a laser adapted to F594 observation. In this study, a 552 nm laser was used (Live mode ON). Set the spectrum of dye emission with the following ranges for each laser: laser 405 (DAPI) from 414 to 483 nm, laser 488 (F488-α-BTX) from 506-531 nm, and laser 552 (NF-M/SV2) from 622-650 nm.
- Collect image stacks of neuromuscular junctions in each experimental group with the same settings: image size 1024 x 1024 pixels (73.7 x 73.7 µm) at 400 Hz sampling rate, Bidirectional X ON, Zoom factor 2.5, Z-step size 0.5 µm in Z-Wide mode.
NOTE: For each NMJ, the number of slices is set to acquire the whole junction. The acquisition settings described above fulfill the Nyquist-Shanon sampling theorem. However, the user can click on the Optimize Format button, present on all recent confocal operating software, to ensure pixel size and Z-step meet the ideal Nyquist sampling rate. This action will avoid over or under-sampled images, which will cause a loss of accuracy in volume measurements. - Save the original file (.lif) or Z-stack images (.tif) in a folder with a name that includes the code name of the slide, the staining type, and the endplate number.
NOTE: Collect sequentially (not simultaneously) the scans using the 488 nm and 552 nm lasers (F488 and F594) to avoid crosstalk of the F488 fluorescence into the F594 channel and vice versa (bleed-through). NB: the beam path can be configured with the Dye Assistant in the microscope software. - Change to the next coded slide and repeat steps 3.1.3-3.1.8 for each NMJ.
- At the end of the session, click on Open in 3D Viewer and choose an NMJ representative of an experimental group to visualize the 3D labeling.
NOTE: This view mode will help to verify that the acquisition parameters were correct. - Close the microscope software, clean the objectives with lens tissues, and then turn off the system.
- Acquisitions by STED microscopy
NOTE: Images were collected with an inverted laser-scanning confocal microscope equipped with Gated STED at 775 nm using a 100x oil immersion objective (HC PL APO CS2 1.4 NA).- For blinded analysis, let a person not involved in the analysis code each slide with a given number. Remain blinded to the experimental groups until the quantification of NMJ parameters is complete for all samples.
- Launch the microscope software in Configuration Mode > machine.xlhw and STED ON (Supplemental Figure 2).
- Click on Open project > New Projects to create a folder to store image acquisitions.
NOTE: Generate a new folder for each slide to limit folder size and prevent computer memory issues. - Place the slide on the microscope stage and view it under wide-field fluorescence illumination using the 488 nm laser to find the observation plane within the sample.
- Search for a NMJ labeled with neurofilament M (NF-M) or SV2 stainings using the 488 nm laser with a spectral detection from 506-531 nm.
- When a NMJ has been identified, click on Activate STED and start acquiring images in a region that contains several junctional folds (Supplemental Figure 3) using the 635 nm laser with a spectral detection from 640-750 nm.
NOTE: Be careful to the saturation look-up table during image acquisition and click the Quick LUT button to avoid overexposure (grey values >255; for 8 bit). - Collect the images of each experimental group with the same settings: image size 2048 x 2048 pixels (38.75 x 38.75 µm) at a 400 Hz sampling rate.
NOTE: The depletion laser (STED) power is set to 65%. - Save the images with a file name that includes the code of the slide.
NOTE: It is possible to click on Optimized XY Format: Set Format to to obtain the best acquisition setting with STED imaging. - Change to the next coded slide and repeat steps 3.2.3-3.2.8. Repeat this procedure for all slides.
- At the end of the STED microscopy session, transfer the image files to another computer and save the original files (.lif) in an external drive or server.
- Turn off the microscope software, clean the objectives with lens tissues, and then turn off the system.
4. Image analysis- confocal microscopy
NOTE: All images were processed with computers using Microsoft Windows 10 professional operating system.
- Launch ImageJ and custom macro to calculate postsynaptic NMJ endplate volume, Maximum Intensity Projection (MIP) area, and relative tortuosity.
- Process NMJ image stacks using NIH ImageJ freeware23, the iGeodesic plugin and the custom macro to obtain NMJ parameter measurements. Launch ImageJ software.
NOTE: The latest version of ImageJ is freely available and can be downloaded24. In order to open proprietary file formats, Bio-Formats Package25 plugin has to be downloaded26 . This step is not necessary in case the operator uses Fiji because the plugin is already installed in the software. The iGeodesic plugin27 to compute tortuosity is also available online28; verify the availability of this plugin in the ImageJ/Fiji version that will be used. The custom-made Macros are also available online29. - Drag and drop the Macro_NMJ_VOL_Marinelloetal.ijm (custom made, Supplementary Coding File 1) to the ImageJ window; the macro will be open in a second window. In this new window, click on Macros > Run Macro.
NOTE: The macro can process both proprietary and TIFF files. Files must fulfill the following criteria: for proprietary file formats, save only one junction (i.e., the stack of images) per file, ordered in a folder; for TIFF images, files must be saved in a folder containing subfolders, each named JunctionX (X corresponds to a NMJ number) with the image stacks of a given junction (RGB TIFF) (Supplemental Figure 4). - Select the native folder containing the Junction subfolders that have to be analyzed and click on Select.
- In the new pop-up menu called Saving Folder, select the storage folder and click on Select.
- In the new pop-up menu called Image Type, select the format of the Z-stack acquisitions.
- Select the RGB channel corresponding to the staining of interest and indicate XY pixel size and Z-step (z). The macro will automatically perform the analysis.
NOTE: In case proprietary file formats are selected, the macro directly reads pixel size and Z-step (z). However, the user still has to indicate the channel of interest (C1, C2, or C3). The macro will provide a DataSheet (.csv) for each junction parameter (endplate volume, MIP area, and tortuosity) in the saving folder. The macro also generates three .TIF files, which correspond to the perimeter of α-BTX staining Drawing_MaxprojX.tif, DrawingJunctionX.tif, and MIP MaxprojX.tif. These TIFF files are generated to verify the quality of the acquisitions and to ensure that image processing has been performed correctly.
Postsynaptic NMJ volume (V): The macro will separate images from a single NMJ and keep the α-bungarotoxin F488 channel corresponding to the postsynaptic endplate. The stack is segmented using Otsu threshold30 on the intermediate slice of the stack. The resulting binary image is 1-pixel dilated, and the Analyze Particles feature is used to measure the endplate area of each detected object. To obtain the postsynaptic NMJ volume, the macro sums all measured endplate areas of the stack and multiplies it by Z-step value (z) in µm.
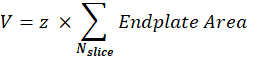
Maximum intensity projection (MIP) endplate area: After the stack is thresholded, the maximum intensity projection (MIP) is obtained using the Z-project ImageJ feature. The Analyze particles feature is then used to quantify the MIP endplate area.
NMJ MIP tortuosity (T): The NMJ tortuosity, which reflects the degree of complexity of the postsynaptic motor endplate including folds and perforations31, is calculated based on each MIP using the following formula, where dObj(AB) is the distance between A and B along the perimeter of the object, and dEuc(AB) is the Euclidian distance between A and B (straight line).
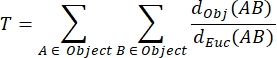
- Set the highest tortuosity value in the wild-type group of each experimental condition to 100%, and normalize all the other values of the experimental condition to this value in order to obtain the relative NMJ tortuosity.
- Process NMJ image stacks using NIH ImageJ freeware23, the iGeodesic plugin and the custom macro to obtain NMJ parameter measurements. Launch ImageJ software.
- Launch ImageJ and custom macro to quantify presynaptic neurofilament accumulation and synaptic vesicle glycoprotein 2 staining.
NOTE: Neurofilament accumulation (here, NF-M) and/or altered distribution of synaptic vesicles (here, SV2) are markers of abnormal axonal transport and/or impaired vesicle trafficking and were previously observed in NMJs of various SMA mouse models32,33,34.- Drag and drop the Macro_NMJ_ACCU_Marinelloetal.ijm (custom made, Supplementary Coding File 2) to the ImageJ window; the macro will open in a second window. In this new window, click on Macros > Run Macro.
NOTE: The macro can process both proprietary file formats and TIFF files. Files must fulfill the criteria indicated in the NOTE below step 4.1.2. - Select the native folder containing the Junction subfolders that have to be analyzed and click on Select.
- In the new pop-up menu called Saving Folder, select the storage folder and click on Select.
- In the new pop-up menu called Image Type, select the format of the Z-stack acquisitions.
- In the Staining Infos pop-up, indicate the presynaptic and postsynaptic label and color and click on OK. For instance, Presynaptic label: SV2 or NF, Presynaptic color: R, Postsynaptic label: BTX, Postsynaptic color: G.
NOTE: For proprietary file formats, labels and corresponding channels (C1, C2, or C3) have to be indicated. - In the Pixel Size pop-up, indicate XY pixel size 0.072 µm and Z step 0.5 µm (z) and click on OK. The macro will automatically perform the analysis.
NOTE: This parameter corresponds to the image size 1024 x 1024 pixels (73.7 x 73.7 µm) selected before confocal microscope acquisitions, and it is correlated to the objective and zoom settings. If the proprietary file formats are selected, the macro directly reads pixel size and Z-step (z). The macro will store, in the saving folder, a DataSheet (.csv) of presynaptic and postsynaptic volumes, a multipage TIFF image of the current detection for each (pre- and postsynaptic) labeling. As indicated above, these TIFF files are generated to check the quality of the acquisitions and to ensure that image processing has been performed correctly.
The macro calculates the volume of axonal neurofilament M staining (NF volume) from the NF-M-F594 channel that colocalizes with α-bungarotoxin-F488 labeling and the volume of NMJ synaptic vesicle glycoprotein 2 staining (SV2 volume) from the SV2-F594 channel that colocalizes with α-BTX-F488 labeling. The NF-M accumulation is quantified by calculating the ratio between NF volume and postsynaptic endplate (α-BTX) volume and NMJ axon terminal occupancy by the ratio of SV2 and α-BTX volumes, as shown below.
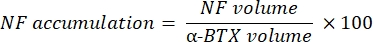
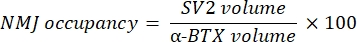
- Drag and drop the Macro_NMJ_ACCU_Marinelloetal.ijm (custom made, Supplementary Coding File 2) to the ImageJ window; the macro will open in a second window. In this new window, click on Macros > Run Macro.
5. Image analysis- STED microscopy
NOTE: Image processing was performed with the offline software of the STED microscope manufacturer.
- Launch the microscope software.
- Open the project by clicking on the Open Project button. Select the project file (.lif) and open it. The images are displayed on the screen along with their names.
- In the Process window: Click on Noise Reduction > Median. At the bottom of the middle window, set Radius to 5.00 and Iteration to 1.00, and untick 3D Filtering.
- Then select the Open Projects tab on the top left of the window and choose an image.
- Click on Apply to validate parameters. A new image called "nameofimage_median001" is created.
NOTE: It is possible to click on Preview prior to Apply to monitor the effect of the median filter, which will enhance the image contrast and smooth the line profiles used for quantification. - Apply the filter to all images as indicated in steps 5.4-5.5.
- In the Open Projects tabs, click on the floppy drive icon to save all the projects, including the newly created filtered images.
NOTE: The next step will be done using the filtered image named "nameofimage_median001". - Calculate the distance between AChR stripes
NOTE: Changes in the morphology of postjunctional folds are often observed in neuromuscular disorders as a sign of NMJ pathology (immaturity or degeneration). The distance (d) between AChR stripes, which are detected by α-bungarotoxin staining, is calculated by generating intensity profiles and quantifying the distance between each maximum intensity peak by drawing a line profile (Supplemental Figure 5).- Using the microscope software, select the Quantify menu on the top of the central window.
- Click on the Tools tab in the top left. Select Intensity in the top left panel and click on the Line profile icon. Set Oversampling to 1, and tick Sort Channels.
- Click on the Open Projects tabs and select the filtered image to be analyzed.
NOTE: It is possible to zoom in on the image by scrolling with the computer mouse. The dynamic range of the image can be modified using the bar on the left side next to the displayed image, which facilitates the visualization of the stripes. - Then, click on the Draw Line icon in the top menu of the right window and trace a line crossing perpendicularly several stripes/junctional folds.
NOTE: The intensity profile is displayed in the central window. - Click on the top of the first peak and move the mouse pointer while keeping the left mouse button pressed until the next maximum peak is reached.
NOTE: The information is shown in the intensity profile, while the distance between the two peaks is displayed under the chart with "dx" denomination. - Click right on the mouse while being in the image of the right window and select Save ROIs. Open the saved ROIs (Regions of Interest) by clicking on Load ROIs.
- Click on the arrow icon in the top left of the right window, click on the ROI and delete it by clicking on the bin icon.
- Repeat this operation as many times as needed from different intensity profiles to obtain the predicted number of AChR stripe distances that will represent the global value in the analyzed muscle.
NOTE: The optimal N value can be calculated in advance based on the estimated difference between groups, α risk, power, and one- or two-tailed test. In the current experimental design, a one-tailed Mann-Whitney test (α risk = 10%; power = 80%) was applied, and the N value was estimated to be at least five AChR stripe distances per NMJ in order to compare the two groups of animals.
- AChR stripe width
NOTE: The stripe width (w) corresponds to the full-width half-maximum (FWHM) of the intensity profile, which is the distance between the points where the α-BTX signal fluorescence value is half of its maximum intensity (Supplemental Figure 5).- Using the microscope software, select the Quantify menu in the central window.
- Click on the Tools tab on the top left side. Select Intensity in the top left panel and click on the Determine FWHM icon. Tick Sort Channels.
NOTE: To optimize the peak detection by the software, Set Threshold and Width were set to 50 and 3, respectively. Adapt these values for each experiment and ask for advice from an experienced imaging scientist. - Click on the Open projects tabs and select the filtered image to be analyzed.
NOTE: It is possible to zoom in on the displayed image in the right window by scrolling with the computer mouse. As indicated above (NOTE following step 5.8.3), the dynamic range of the image can be modified for optimal stripe visualization. - Next, click on the Draw Rectangle icon in the top menu of the right window. Select a stripe that is either horizontal or vertical and draw a rectangle perpendicularly to the stripe. A profile appears in the central window.
- Click on either Vertical or Horizontal of the Average Projection menu located in the left panel, depending on whether the stripe orientation is vertical or horizontal.
- Click on Statistics in the central window and read the FWHM value.
- Make a right-click with the computer mouse on the image displayed in the right window and select Save ROIs.
NOTE: Open the saved ROIs by clicking on Load ROIs. - Click on the arrow icon in the top left of the right window, click on the ROI and delete it by clicking on the bin icon.
- Repeat this operation as many times as needed from different rectangle ROIs until obtaining the predicted number of AChR stripe widths, which will be representative of the global value in the analyzed muscle.
6. Experimental design and statistical tests
- Perform statistical analyses using specific software.
NOTE: Data were collected from N ≥ 3 biological replicates and at least 20 NMJs per genotype for confocal microscope imaging, and N ≥ 5 biological replicates and N = 5 NMJs per genotype for STED imaging, in each experimental group. Significance was assessed by unpaired Mann-Whitney test (nonparametric), and p-values are indicated in the corresponding figure legends.
Results
In order to facilitate the morphological analysis of neuromuscular junctions at the pre- and postsynaptic level in a reproducible manner, a workflow was developed from muscle harvesting to imaging and quantification using the microscope software and ImageJ custom macros (Figure 1). To exemplify the utility of this protocol, the morphology of NMJs in two mouse models of genetic disorders, Smn2B/- and ColQDex2/Dex2 mice affected by spinal muscular atrophy (SMA) and a congenital myasthenic syndrome (CMS) form, respectively, were evaluated and data were compared to age-matched control littermates.
The NMJ structure was assessed from tibialis anterior and gastrocnemius muscles of 3- and 6-week-old Smn2B/- (C57Bl/6 background) and ColQDex2/Dex2 (B6D2F1/J background) mice, respectively, when signs of the disease are already present in these animals. At 3 weeks of age, Smn2B/- mice show signs of delayed skeletal muscle development and denervation, such as NMJ atrophy and loss35,36. CMS mice have a primary pathology in NMJs and manifest a reduction in body weight from the first week of life and marked muscle weakness20 (data not shown). As shown in Figure 2A, the postsynaptic motor endplate labeled with fluorescent α-bungarotoxin appeared smaller and/or fragmented in mutants of the two mouse lines by confocal microscopy. Quantification of NMJ Z-stacks using this customized ImageJ macros revealed marked decreases in endplate volume, maximum intensity projection (MIP), and relative tortuosity in both SMA and CMS mice compared to controls, as signs of NMJ maturation defects32 (Figure 2B-D). Postsynaptic endplate volume and MIP were decreased in diseased animals (fold-change of 2.7 and 2.0 for volume, and 2.5 and 2.0 for MIP, in Smn2B/- and ColQDex2/Dex2 mice, respectively). The relative tortuosity was also smaller in SMN and ColQ deficient muscles than WT (16.97% ± 1.33% in SMA versus 48.84% ± 5.90% WT mice, and 13.29% ± 2.79% in CMS versus 30.20% ± 4.44% control mice). In addition, the quantification of the distribution of presynaptic axon terminal branches using the ImageJ custom macro revealed an altered pattern in neurofilament M distribution in the two animal models, with increased immunolabelling (84.65% ± 0.32% versus 16.57% ± 2.03% and 23.64% ± 2.78% versus 18.77% ± 1.73% in Smn2B/- and ColQDex2/Dex2 mice compared to controls, respectively) (Figure 3A-D). By SV2 staining, a 43% reduction in the occupancy ratio, i.e., percent of AChR-containing regions with adjacent nerve terminal active zones, was also observed in Smn2B/- mice (49.36% ± 3.76% in SMA versus 85.69% ± 2.34% WT mice) (Figure 3E,F). This NMJ parameter was also calculated in GA of ColQDex2/Dex2 mutants, but no statistically significant difference was found compared to control littermates (data not shown).
We further analyzed postsynaptic membrane characteristics by quantifying the distance between junctional folds and the width of AChR stripes, which are located at the crest of these folds, in ColQ-deficient muscle using super-resolution stimulated emission depletion (STED) microscopy. As shown in Figure 4, the aspect of these structures can be clearly visualized by fluorescent α-bungarotoxin labeling and intensity profile analysis. We evaluated these NMJ parameters and found an increase in the junctional fold distance (d) and width (w) of AChR stripes in the gastrocnemius muscle of mutants (358.3 nm ± 11.97 nm and 320.8 nm ± 10.90 nm for the distance, and 216.9 nm ± 10.51 nm and 186.3 nm ± 7.015 nm for the width, in ColQDex2/Dex2 as compared to wild-type mice, respectively, p < 0.05) (Figure 4C,D).
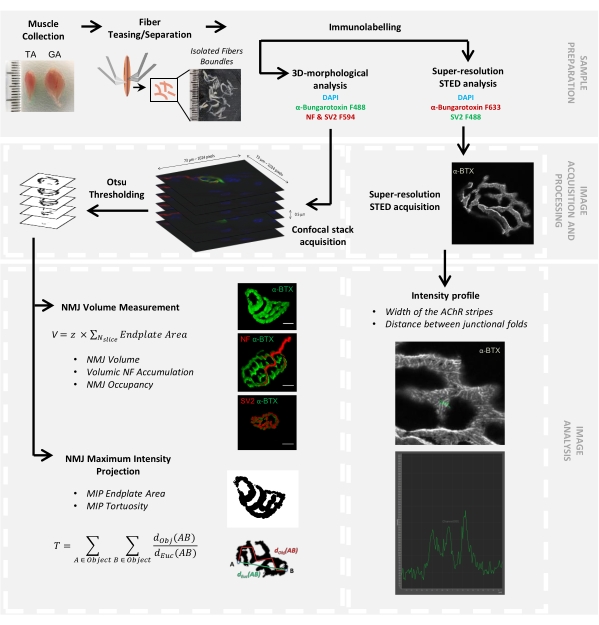
Figure 1: Flowchart of the video protocol for 3D multiscale NMJ characterization by confocal and STED microscopy. Tibialis anterior (TA) and gastrocnemius (GA) muscles were collected from mice, and muscle fibers were teased before labeling with α-bungarotoxin-F488 or α-bungarotoxin-F633, DAPI, primary antibodies directed against neurofilament M (NF-M) and synaptic vesicle glycoprotein 2 (SV2), and fluorophore (F488 or F594)-conjugated secondary antibodies. Image stacks were acquired by confocal microscopy and processed to measure postsynaptic NMJ volume, presynaptic NF-M accumulation, NMJ axon terminal occupancy, postsynaptic maximum intensity projection (MIP) endplate area, and tortuosity (dObj(AB) is the distance between A and B along the perimeter of the object (red line), whereas dEuc(AB) is the Euclidian distance between A and B (green line)). For STED microscopy analysis, the width of acetylcholine receptor (AChR) stripes and the distance between junctional folds were quantified from intensity profiles of α-BTX-F633 staining. Please click here to view a larger version of this figure.
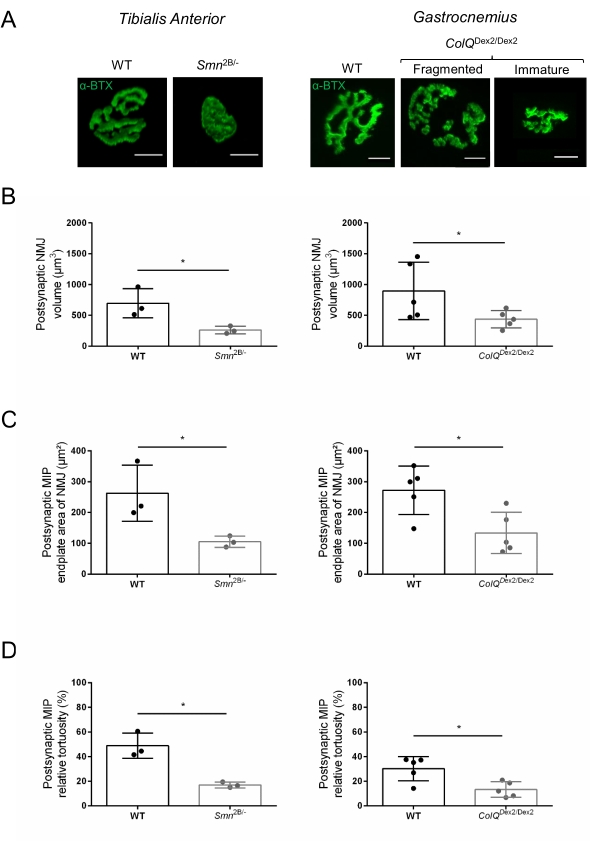
Figure 2: Multi-parameter postsynaptic NMJ characterization in mouse models of spinal muscular atrophy (SMA) and ColQ-related congenital myasthenic syndrome (CMS). (A) Representative images of postsynaptic motor endplates from TA and GA muscles labeled with α-bungarotoxin-F488 (α-BTX). (B) Quantification of NMJ postsynaptic endplate volume, (C) maximal intensity projection (MIP) area and (D) relative tortuosity in TA of 3 week-old wild-type (WT) and Smn2B/- mice (left graphs, N = 3 animals per genotype, n = 37 and n = 56 NMJs, respectively) and 6 week-old WT and ColQDex2/Dex2 mice (right graphs, N = 5 mice per genotype, n = 89 and n = 97 NMJs, respectively). Data are expressed as the mean per mouse (dot) ± SEM. Differences between groups were analyzed by Mann-Whitney test (* p < 0.05). Scale bar is 10 µm. Please click here to view a larger version of this figure.
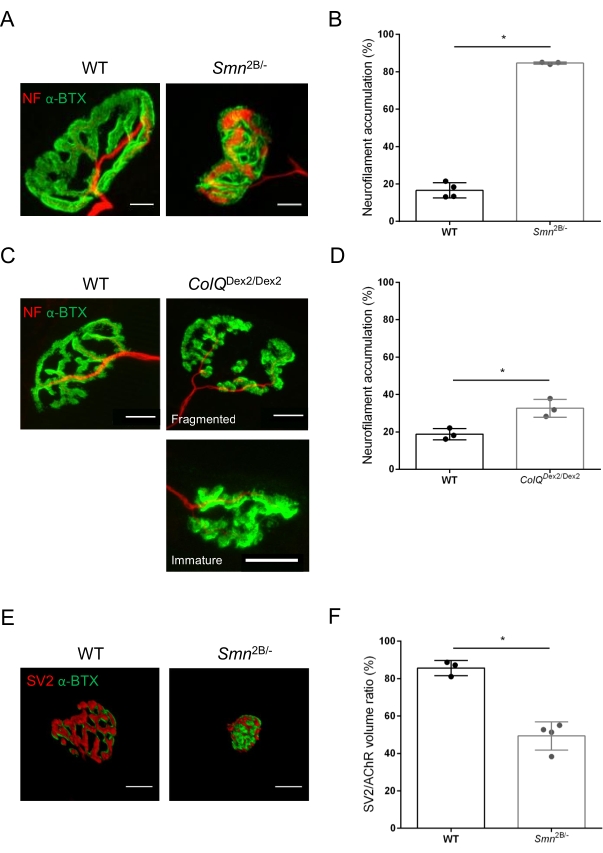
Figure 3: Morphometric analysis of presynaptic axon terminal distribution in muscles of WT and mutant mice. NMJ innervation pattern in tibialis anterior (TA) and gastrocnemius (GA) muscles of wild-type, SMA and ColQ-related CMS mice. (A, B) Representative neuromuscular junctions from TA of WT and Smn2B/- mice at 21 days of age labeled with antibodies against neurofilament M (NF-M, red) and α-bungarotoxin-F488 (α-BTX, green) (A), and results from quantitative analysis of neurofilament accumulation (B); (C, D) Representative neuromuscular junctions from GA of 6 week-old WT and ColQDex2/Dex2 mice labeled with antibodies against neurofilament M (NF-M, red) and α-bungarotoxin-F488 (α-BTX, green), showing fragmented and immature postsynaptic endplates (C), and results of neurofilament accumulation in the two groups of animals (D). N= 4 (n = 34 NMJs) (B) and N = 3 (n = 54 NMJs) (D) WT animals, and N=3 (n = 36 NMJs) Smn2B/- and N = 3 (n = 55 NMJs) ColQDex2/Dex2 mice were analyzed in the experiments (B, D). (E, F) Representative images of axon terminal occupancy in NMJs from TA of 3 week-old WT and Smn2B/- mice labeled with antibodies against synaptic vesicle glycoprotein 2 (SV2, red) and α-bungarotoxin-F488 (α-BTX, green) (E), and results of NMJ occupancy (SV2/AChR volume ratio) (F). Muscles from N = 3 (n = 50 NMJs) wild-type and N = 4 (n = 62 NMJs) Smn2B/- mice were analyzed. Data are expressed as the mean value per mouse (dot) ± SEM. Differences between groups were analyzed by Mann-Whitney test (* p < 0.05). Scale bars are 20 µm. Please click here to view a larger version of this figure.
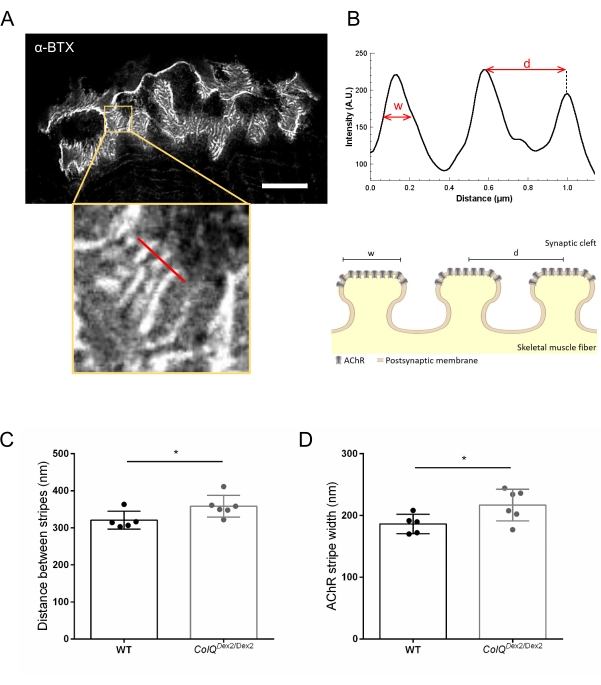
Figure 4: STED imaging of NMJ postsynaptic endplates. (A) Representative STED image of a NMJ labeled with α-bungarotoxin-F633 (α-BTX) from gastrocnemius of a 6 week-old wild-type mouse showing postjunctional AChR stripes (scale bar is 5 µm). (B) Higher magnification of a region with AChR stripes (bottom panel) that was used to generate the intensity profile. The width (w) of AChR stripes and the distance between two adjacent stripes (d) of this region were quantified and presented in the bar graph. Schematic representation of the postsynaptic endplate to illustrate AChR stripe width (w) and distance (d). These parameters, (C) AChR stripe distance and (D) width, were measured in ColQDex2/Dex2 mice and control littermates at 6 weeks of age. NMJs from 5 WT (total n = 29 NMJs) and 6 ColQDex2/Dex2 (total n = 43 NMJs) animals were analyzed blindly. Data are expressed as the mean per mouse (dot) ± SEM. Statistical differences between groups were analyzed by using the Mann-Whitney test (* p < 0.05). Please click here to view a larger version of this figure.
Supplemental Figure 1: Launch of LAS X software and parameters for confocal acquisitions. The various steps to acquire confocal images are described in sections 3.1.2 to 3.1.7 of the protocol. For each NMJ stack acquisition, a project is opened (step 3.1.4) and the parameters of image size, acquisition speed, X, Y and Z axes are selected (step 3.1.7), with each sequential scan indicated (Seq.1, laser 405 for DAPI; Seq.2, laser 488 for α-BTX-F488; and Seq.3, laser 552 for F594 conjugated secondary antibodies). Please click here to download this File.
Supplemental Figure 2: Launch of LAS X software and parameters for STED acquisitions. The steps to acquire STED images are described in sections 3.2.2 to 3.2.8 of the protocol. The microscope is launched in configuration mode STED ON (step 3.2.2), and a project is opened (step 3.2.3). The parameters for image acquisition (step 3.2.7) (image size, acquisition speed, Zoom factor, X axis), with each sequential scan are indicated (Seq.1 for α-BTX-F633; Seq.2 for F488 conjugated secondary antibodies). Please click here to download this File.
Supplemental Figure 3: Images of α-BTX-stained junctional folds obtained by STED microscopy. Image examples of a postsynaptic endplate labeled with α-BTX-F633 from a 6 week-old wild-type mouse that were acquired with either a correct (left) or incorrect focus (right). Please click here to download this File.
Supplemental Figure 4: Windows pop-ups to describe the input and output data obtained by the custom ImageJ macros. Input data examples (.tif and .lif files) of NMJ images are shown on the left column. The output data from the macros (right column) are saved in folders (Save_Volume, Save_Accu) that contain images of the junction (.tif) and datasheets containing the results (.csv files). Please click here to download this File.
Supplemental Figure 5: AChR stripe distance and width analysis from a STED acquisition using the LAS X software. The steps to analyze NMJ STED images are described in section 5 of the protocol. A) Image of a labeled postsynaptic endplate region containing AChR stripes. The region of interest for stripe analysis is selected by drawing a perpendicular line (green line, for stripe distance), or a perpendicular rectangle (purple rectangle, for stripe width). (B, C) Intensity profiles of the selected regions and measurements to calculate the distance between AChR stripes (B) and the AChR stripe width (C) are shown. Please click here to download this File.
Supplementary Coding File 1: Macro_NMJ_VOL_Marinelloetal. ImageJ custom macro to extract NMJ parameter measurements (NMJ volume, MIP endplate area, and NMJ tortuosity). Please click here to download this File.
Supplementary Coding File 2: Macro_NMJ_ACCU_Marinelloetal. ImageJ custom macro to extract NF-M accumulation and SV2 staining. Please click here to download this File.
Discussion
The described video protocol provides a detailed method to quantify the 3D structure of neuromuscular junctions by combining confocal and STED microscopy that can be used to characterize pathological changes at the pre- and postsynaptic levels. The high resolution of STED microscopy allows visualization and morphometric analysis of nanostructures that are not identifiable by conventional confocal imaging. This procedure enabled us to measure structural alterations of NMJs in two appendicular muscles, tibialis anterior and gastrocnemius, of SMA and ColQ-related CMS mice.
To obtain reliable results with this technique, it is critical to dissect and tease muscles properly, paying particular attention to the fascia surrounding the muscle and the applied strength to separate muscle bundles; otherwise, the innervation pattern could be disrupted impeding proper presynaptic NMJ assessment. Although detailed information is provided to analyze NMJs from TA and GA, in principle, this protocol could be adapted to other muscles, including flat muscles, such as the diaphragm or transverse abdominis37, which do not require the teasing step. Tissue fixation is also crucial to ensure good quality staining; therefore, it is recommended to use high-quality PFA at an appropriate volume (15-20 times that of the muscle). In addition, the exposure time to the fixative is an important step because artifacts, such as shrinkage and clumping, may appear due to over-fixation and influence NMJ features. Given the size of the samples and the penetration rate of the paraformaldehyde solution in tissues38, a fixation time of 18-24 h is recommended for this type of muscle. In case the staining step is planned more than a week after tissue harvesting, it is suggested to keep PFA-fixed muscles in PBS supplemented with sodium azide at 4 °C to prevent bacterial proliferation.
This protocol presents an approach using α-BTX-F488 for confocal and α-BTX-F633 for STED imaging. These fluorophores were chosen to fit with the described experimental design but can be modified according to the available equipment and materials. For instance, α-BTX F488 labeling can be selected when using a STED CW 592 nm laser for image acquisitions and quantification. However, it appears that the configuration that was applied in the present study (pulsed excitation gated STED, 775 nm depletion) exhibits higher performance and better resolution than other approaches, such as continuous wave STED39, making it more suitable for the current application. It is also important to select carefully the laser power settings, especially for STED (both excitation and depletion), since the characteristics of an intensity profile cannot be measured in case of saturation, and therefore any saturated signal in a NMJ image could jeopardize the whole analysis.
This detailed workflow, including image acquisitions and analysis using microscope software and ImageJ macros, was developed to facilitate autonomous NMJ morphometric analysis by confocal and STED microscopy from a single muscle. Previously described workflows for NMJ confocal analysis, such as NMJ-morph2 or NMJ-Analyser14, paved the way for the design of semi-automatic methods that facilitate morphological analysis of NMJs and comparative studies. NMJ-morph (and its updated version aNMJ-morph15) is a free ImageJ-based platform that uses the maximal intensity projection to measure 21 morphological features, and NMJ-Analyser uses a script developed in Python that generates 29 relevant parameters from the entire 3D NMJ structure. Manual thresholding is the only step during image processing in these two methods that require user analysis. This integrated protocol details steps for tissue preparation, 3D confocal image acquisitions, and ImageJ-based processing of NMJs from entire skeletal muscles and provides a simplified overview of five important parameters of the postsynaptic (volume, maximal projection area, and tortuosity) and presynaptic (axon terminal occupancy and neurofilament accumulation) endplates. An additional parameter of biological relevance, the AChR organization pattern of the postsynaptic junctional folds, was incorporated for morphometric analysis at the nanoscale level by super-resolution STED microscopy (resolution 20-30 nm)40. Interestingly, tissue preparation for STED imaging is simpler than other methods used for NMJ ultrastructural studies, such as conventional transmission electron microscopy (TEM)9, which is a rather complex and time-consuming procedure that requires a skilled manipulator in order to obtain ultrathin sections of the appropriate muscle region. In addition, quantitative data from multiple junctional folds can be obtained automatically by using the STED-associated software.
This protocol was applied to illustrate previously known NMJs defects in SMN and ColQ deficient muscles20,36,41,42. Common changes were found in the two mouse models by confocal microscopy, such as decreased postsynaptic endplate volume, MIP area, and relative tortuosity, and increased neurofilament accumulation, whereas some more specific findings (decreased NMJ occupancy), were observed only in SMA mice, as an indicator of impaired vesicles trafficking36. Finally, an increase in AChR stripe distance and width were detected in ColQ-KO by STED analysis, which are signs of ultrastructural defects in the postsynaptic junctional folds, as previously observed by TEM20. Importantly, this protocol may help in a more in-depth morphological characterization of neuromuscular junctions during development, maintenance, and under various pathological conditions.
Disclosures
The authors declare no conflicts of interest related to this work.
Acknowledgements
We thank Genethon's "Imaging and Cytometry Core Facility", as well as the histology service, which are supported in part by equipment funds from the Region Ile-de-France, the Conseil General de l'Essonne, the Genopole Recherche of Evry, the University of Evry Val d'Essonne and the INSERM, France. We are also grateful to Dr. Rashmi Kothary for providing the Smn2B/2B mouse line (University of Ottawa, Canada) and Dr. Eric Krejci for the ColQDex2/+ mouse line (unpublished, University of Paris, France). We thank Guillaume Corre for his support in statistical analysis. The 2H3 (developed by Jessel, T.M. and Dodd, J.) and SV2 (developed by Buckley, K.M.) monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank (DSHB), created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was supported by the Association Française contre les Myopathies (AFM-Telethon), the INSERM and the University of Evry Val d'Essonne.
Materials
| Name | Company | Catalog Number | Comments |
| Buffers and Reagents | |||
| Alexa Fluor 488 goat anti-mouse IgG (F488) | Life Technologies, Thermofisher | A-11001 | |
| Alexa Fluor 488 α-bungarotoxin (F488-a-BTX) | Life Technologies, Thermofisher | B13422 | |
| Alexa Fluor 594 goat anti-mouse IgG (F594) | Life Technologies, Thermofisher | A-11032 | |
| ATTO-633 α-bungarotoxin (F633-a-BTX) | Alomone Labs | B-100-FR | |
| Bovine serum albumin (BSA) | Sigma | A2153 | |
| DAPI Fluoromount-G | Southern Biotech | 00-4959-52 | |
| DPBS | Gibco, Invitrogen | 14190-169 | |
| Ethanol Absolute | VWR | 20821.296 | |
| Immersion Oil, n = 1.518 | THORLABS | MOIL-10LF | Low autofluorescence |
| Neurofilament (NF-M) antibody | DSHB | AB_531793 | |
| Paraformaldehyde (PFA) | MERCK | 1.04005 | |
| Synaptic vesicle glycoprotein 2 (SV2) antibody | DSHB | AB_2315387 | |
| Triton X-100 | Sigma | T8787 | |
| Materials | |||
| Alnico Button cylindrical magnets | Farnell France | E822 | diameter of 19.1 mm with maximal pull of 1.9 Kg |
| 63x 1.4 NA magnitude oil immersion HCX Plan Apo CS objective | Leica Microsystems | ||
| 100x 1.4 NA HC PL APPO CS2 Objective | Zeiss | ||
| Curved thin forceps-Moria iris forceps | Fine Science Tools | 11370-31 | |
| Extra thin scissors - Vannas-Tübingen Spring Scissors | Fine Science Tools | 15-003-08 | |
| Fine serrated forceps | Euronexia | P-95-AA | |
| Gel loading tip round 1-200 µL | COSTAR | 4853 | |
| Leica laser-scanning confocal microscope TCS SP8 | Leica Microsystems | ||
| Leica Laser-scanning confocal microscope TCS SP8 Gated STED 775 nm | Leica Microsystems | ||
| Lens Cleaning Tissue | Whatman (GE Healthcare) | 2105-841 | |
| Medium serrated forceps | Euronexia | P-95-AB | |
| Microscope cover glasses 24x50 nm No 1.5H 170±5 µm | Marienfield | 107222 | High precision |
| Nunclon delta surface (12-well plates) | Thermo Scientific | 150628 | |
| Nunclon delta surface (24-well plates) | Thermo Scientific | 142475 | |
| Safeshield scalpel | Feather | 02.001.40.023 | |
| Sharp-blunt scissors - fine Scissors - Martensitic Stainless Steel | Fine Science Tools | 14094-11 | |
| Superfrost plus slides | Thermo Scientific | J1800AMNZ | |
| Software | |||
| GraphPad | Prism, San Diego (US) | Release N°6.07 | Statistical software |
| ImageJ software | National Institutes of Health | Release N° 1.53f | |
| Leica Application Suite X software | Leica Microsystems | Release N°3.7.2.2283 | Free microscope software available at https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/downloads/ |
References
- Slater, C. R. Postnatal maturation of nerve-muscle junctions in hindlimb muscles of the mouse. Developmental Biology. 94 (1), 11-22 (1982).
- Jones, R. A., et al. NMJ-morph reveals principal components of synaptic morphology influencing structure-function relationships at the neuromuscular junction. Open Biology. 6 (12), (2016).
- Willadt, S., Nash, M., Slater, C. Age-related changes in the structure and function of mammalian neuromuscular junctions. Annals of the New York Academy of Sciences. 1412, 41-53 (2018).
- Boehm, I., et al. Comparative anatomy of the mammalian neuromuscular junction. Journal of Anatomy. 237 (5), 827-836 (2020).
- Nishimune, H., Shigemoto, K. Practical anatomy of the neuromuscular junction in health and disease. Neurologic Clinics. 36 (2), 231-240 (2018).
- Moloney, E. B., de Winter, F., Verhaagen, J. ALS as a distal axonopathy: molecular mechanisms affecting neuromuscular junction stability in the presymptomatic stages of the disease. Frontiers in Neuroscience. 8, (2014).
- Lovering, R. M., Iyer, S. R., Edwards, B., Davies, K. E. Alterations of neuromuscular junctions in Duchenne muscular dystrophy. Neuroscience Letters. 737, 135304(2020).
- Koneczny, I., Herbst, R. Myasthenia Gravis: Pathogenic effects of autoantibodies on neuromuscular architecture. Cells. 8 (7), 671(2019).
- Dowling, J. J., et al. Myotubular myopathy and the neuromuscular junction: a novel therapeutic approach from mouse models. Disease Models & Mechanisms. 5 (6), 852-859 (2012).
- Gibbs, E. M., et al. Neuromuscular junction abnormalities in DNM2-related centronuclear myopathy. Journal of Molecular Medicine. 91 (6), 727-737 (2013).
- Swoboda, K. J., et al. Natural history of denervation in SMA: Relation to age, SMN2 copy number, and function. Annals of Neurology. 57 (5), 704-712 (2005).
- Rodríguez Cruz, P. M., Palace, J., Beeson, D. The neuromuscular junction and wide heterogeneity of congenital myasthenic syndromes. International Journal of Molecular Sciences. 19 (6), 1677(2018).
- Tse, N., et al. The neuromuscular junction: Measuring synapse size, fragmentation and changes in synaptic protein density using confocal fluorescence microscopy. Journal of Visualized Experiments: JoVE. (94), e52220(2014).
- Mejia Maza, A., et al. NMJ-Analyser identifies subtle early changes in mouse models of neuromuscular disease. Scientific Reports. 11 (1), 12251(2021).
- Minty, G., et al. aNMJ-morph: a simple macro for rapid analysis of neuromuscular junction morphology. Royal Society Open Science. 7 (4), 200128(2020).
- Modla, S., Mendonca, J., Czymmek, K. J., Akins, R. E. Identification of neuromuscular junctions by correlative confocal and transmission electron microscopy. Journal of Neuroscience Methods. 191 (2), 158-165 (2010).
- Kittel, R. J., et al. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 312 (5776), 1051-1054 (2006).
- York, A. L., Zheng, J. Q. Super-resolution microscopy reveals a nanoscale organization of acetylcholine receptors for trans-synaptic alignment at neuromuscular synapses. eNeuro. 4 (4), (2017).
- Bowerman, M., Murray, L. M., Beauvais, A., Pinheiro, B., Kothary, R. A critical smn threshold in mice dictates onset of an intermediate spinal muscular atrophy phenotype associated with a distinct neuromuscular junction pathology. Neuromuscular Disorders. 22 (3), 263-276 (2012).
- Feng, G., Krejci, E., Molgo, J., Cunningham, J. M., Massoulié, J., Sanes, J. R. Genetic analysis of collagen Q: Roles in acetylcholinesterase and butyrylcholinesterase assembly and in synaptic structure and function. Journal of Cell Biology. 144 (6), 1349-1360 (1999).
- Sigoillot, S. M., et al. Neuromuscular junction immaturity and muscle atrophy are hallmarks of the ColQ-deficient mouse, a model of congenital myasthenic syndrome with acetylcholinesterase deficiency. The FASEB Journal. 30 (6), 2382-2399 (2016).
- Vanhaesebrouck, A. E., Beeson, D. The congenital myasthenic syndromes: expanding genetic and phenotypic spectrums and refining treatment strategies. Current Opinion in Neurology. 32 (5), 696-703 (2019).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH image to ImageJ: 25 years of image analysis. Nature Methods. 9 (7), 671-675 (2012).
- ImageJ. , Available from: http://imagej.nih.gov/ij (2021).
- Linkert, M., et al. Metadata matters: access to image data in the real world. Journal of Cell Biology. 189 (5), 777-782 (2010).
- Bio-Formats. , Available from: http://www.openmicroscopy.org/bio-formats/downloads/ (2021).
- Legland, D., Beaugrand, J. Automated clustering of lignocellulosic fibres based on morphometric features and using clustering of variables. Industrial Crops and Products. 45, Supplement C 253-261 (2013).
- ImageJ documentation. , Available from: http://imagejdocu.tudor.lu/plugin/analysis/geodesic_distances/start (2021).
- GitHUb. , Available from: https://github.com/Genethon/ImCy (2021).
- Otsu, N. A Threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics. 9 (1), 62-66 (1979).
- Sanes, J. R., Lichtman, J. W. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nature Reviews Neuroscience. 2 (11), 791-805 (2001).
- Kong, L., et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. The Journal of Neuroscience. 29 (3), 842-851 (2009).
- Cifuentes-Diaz, C., et al. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Human Molecular Genetics. 11 (12), 1439-1447 (2002).
- Murray, L. M., Comley, L. H., Thomson, D., Parkinson, N., Talbot, K., Gillingwater, T. H. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Human Molecular Genetics. 17 (7), 949-962 (2008).
- Boyer, J. G., et al. Myogenic program dysregulation is contributory to disease pathogenesis in spinal muscular atrophy. Human Molecular Genetics. 23 (16), 4249-4259 (2014).
- Ling, K. K. Y., Gibbs, R. M., Feng, Z., Ko, C. -P. Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Human Molecular Genetics. 21 (1), 185-195 (2012).
- Murray, L., Gillingwater, T. H., Kothary, R. Dissection of the transversus abdominis muscle for whole-mount neuromuscular junction analysis. Journal of Visualized Experiments: JoVE. (83), e51162(2014).
- Baker, J. R. Principles of biological microtechnique; a study of fixation and dyeing. , Wiley. London Methuen; New York. at http://archive.org/details/principlesofbiol01bake (1958).
- Vicidomini, G., et al. STED Nanoscopy with time-gated detection: Theoretical and experimental aspects. PLoS ONE. 8 (1), 054421(2013).
- Badawi, Y., Nishimune, H. Super-resolution microscopy for analyzing neuromuscular junctions and synapses. Neuroscience Letters. 715, 134644(2020).
- Thomson, S. R., et al. Morphological characteristics of motor neurons do not determine their relative susceptibility to degeneration in a mouse model of severe spinal muscular atrophy. PLoS ONE. 7 (12), 052605(2012).
- McMacken, G. M., et al. Salbutamol modifies the neuromuscular junction in a mouse model of ColQ myasthenic syndrome. Human Molecular Genetics. 28 (14), 2339-2351 (2019).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved