A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Rectal Organoid Morphology Analysis (ROMA): A Diagnostic Assay in Cystic Fibrosis
In This Article
Summary
This protocol describes rectal organoid morphology analysis (ROMA), a novel diagnostic assay for cystic fibrosis (CF). Morphological characteristics, namely the roundness (circularity index, CI) and the presence of a lumen (intensity ratio, IR), are a measure of CFTR function. Analysis of 189 subjects showed perfect discrimination between CF and non-CF.
Abstract
Diagnosis of cystic fibrosis (CF) is not always straightforward, especially when sweat chloride concentration is intermediate and/or less than two disease-causing CFTR mutations can be identified. Physiological CFTR assays (nasal potential difference, intestinal current measurement) have been included in the diagnostic algorithm but are not always readily available or feasible (e.g., in infants). Rectal organoids are 3D structures that grow from stem cells isolated from crypts of a rectal biopsy when cultured under specific conditions. Organoids from non-CF subjects have a round shape and a fluid-filled lumen, as CFTR-mediated chloride transport drives water into the lumen. Organoids with defective CFTR function do not swell, retaining an irregular shape and having no visible lumen. Differences in morphology between CF and non-CF organoids are quantified in the 'Rectal Organoid Morphology Analysis' (ROMA) as a novel CFTR physiological assay. For the ROMA assay, organoids are plated in 96-well plates, stained with calcein, and imaged in a confocal microscope. Morphological differences are quantified using two indexes: The circularity index (CI) quantifies the roundness of organoids, and the intensity ratio (IR) is a measure of the presence of a central lumen. Non-CF organoids have a high CI and low IR compared to CF organoids. ROMA indexes perfectly discriminated 167 subjects with CF from 22 subjects without CF, making ROMA an appealing physiological CFTR assay to aid in CF diagnosis. Rectal biopsies can be routinely performed at all ages in most hospitals and tissue can be sent to a central lab for organoid culture and ROMA. In the future, ROMA might also be applied to test the efficacy of CFTR modulators in vitro. The aim of the present report is to fully explain the methods used for ROMA, to allow replication in other labs.
Introduction
Cystic fibrosis (CF) is an autosomal recessive disease caused by mutations in the CF transmembrane conductance regulator (CFTR) gene. The CFTR protein is a chloride and bicarbonate channel, ensuring hydration of several epithelia1. CF is a high-burden, life-shortening, multi-system disease, manifesting primarily as a respiratory disease, but also affecting the gastrointestinal tract, pancreas, liver, and reproductive tract2.
Disease-causing CFTR mutations lead to a decrease in the amount or function of CFTR, in turn causing mucus dehydration. More than 2,000 variants in the CFTR gene have been described3, of which only 466 have been thoroughly characterized4.
A diagnosis of CF can be made when either sweat chloride concentration (SCC) is above the threshold of 60 mmol/L or when two disease-causing CFTR mutations (according to the CFTR2 database) are identified4,5. In subjects with only intermediately elevated (30-60 mmol/L) SCC, which occurs in about 4%-5% of sweat tests6, and CFTR mutations of varying or unknown clinical consequence, the diagnosis cannot be confirmed nor ruled out, even when they have CF compatible symptoms or a positive neonatal screening test. For these cases, second-line physiological CFTR assays (nasal potential difference (NPD) and intestinal current measurements (ICM)) have been included in the diagnostic algorithm. These tests are not readily available at most centers nor feasible at all ages, especially in infants5.
Rectal organoids are 3D structures grown from Lgr5(+) adult intestinal stem cells from intestinal crypts obtained through rectal biopsy7. Organoids are being used increasingly in biomedical research, such as testing modulator treatment in CF8. A viable biopsy can be obtained by either suction or forceps biopsy, a procedure that causes only minimal discomfort and is safe even in infants, with low complication rates9. The crypts isolated from the rectal biopsies are enriched in stem cells, and under specific culturing conditions, these self-organize into rectal organoids. The morphology of these organoids is determined by the expression and function of CFTR, located at the apical membrane of epithelial cells. Functional CFTR allows chloride and water to enter the organoid lumen, thereby inducing swelling of non-CF organoids. CF organoids do not swell and have no visible lumen10,11.
Rectal organoid morphology analysis (ROMA) allows the discrimination between CF and non-CF organoids based on these differences in organoid morphology. Non-CF organoids are more round and have a visible lumen, while the opposite is true for CF organoids. For this assay, patient-specific organoids are plated in 32 wells of a 96-well plate. After 1 day of growing, the organoids are stained with calcein green and imaged in a confocal microscope. The non-CF organoids show a more circular shape and a less fluorescent central part, as the lumen contains fluid and calcein stains only cells. These differences in morphology are quantified using two ROMA indexes: the circularity index (CI) quantifies the roundness of organoids, while the intensity ratio (IR) is a measure of the presence or absence of a central lumen. In this report, we describe in detail the protocol to obtain these discriminative indexes, to allow replication of the technique.
Protocol
For all procedures involving human tissue, approval by the Ethics Committee Research UZ/KU Leuven (EC research) was acquired. All research was performed with informed consent and/or assent from parents, representatives, and/or patients.
NOTE: All procedures involving rectal biopsies and organoids should be performed in a laminar flow to protect the researcher from any biological hazard and to minimize the risk of contamination of the cultures. As for any lab procedure, researchers should at all times wear lab coats, gloves, and safety goggles to manipulate samples.
1. Rectal biopsy, isolation of adult stem cells from crypts, and organoid culture
- For this initial part of the protocol, including media production and usage, organoid culture, splitting, expansion, and freezing for biobanking, follow the two previously published protocols8,12.
- In short, the following steps have to be taken:
- Take three to four rectal biopsies with a forceps or suction device and collect in a sterile container (e.g., 1.5 mL microcentrifuge tube) with the Ad-DF+++ medium as described in the protocol mentioned above.
- Transport biopsies to a central lab on ice or at 4 °C. Transport to other centers is possible, and quality is not significantly affected even when transport takes up to 48 h. If transport is expected to take over 6 h, use a 15 mL conical tube with 6 mL of Ad-DF+++ medium instead of a 1.5 mL microcentrifuge tube.
- Wash the biopsies in cold PBS until the supernatant is clear to remove the debris and non-epithelial tissues such as fat tissue.
- Incubate the biopsies with EDTA (final concentration 10 mM) to detach the crypts. Plate the crypts in a basement membrane matrix. Add broad-spectrum antibiotics (gentamicin 50 µg/mL and vancomycin 50 µg/mL) to the medium in the first week of culturing to prevent bacterial contamination.
- When the crypts have budded and are closed and proliferated, perform mechanical splitting, usually after 7 days.
- Split the resulting organoids approximately every 7 days. This way, expand the culture, freeze backup samples in a biobank, or use the organoids for assays.
2. Organoid plating for ROMA (day 1)
- Mechanically split the organoids for plating
- Collect organoids from three well-grown wells from a 24-well plate by washing twice with cold Ad-DF+++ medium, collect them in a 1.5 mL microcentrifuge tube, and assess them under the microscope12.
- Ensure that organoids are viable and of high quality, which can usually be achieved after 5-7 days of growth after being split previously (Figure 1).
- Mechanically split the organoids according to the aforementioned protocol8,12. Repeat until the majority of the organoids, as assessed using the brightfield microscope, are small enough compared to the initial observation.
- Collect the smaller organoids, usually corresponding to the collection of about the top 4/5 of the organoid-medium solution in a new microcentrifuge tube, as big organoids sink to the bottom of the tube due to gravity and small organoids stay suspended in the upper part of the medium column.
- Centrifuge the sample of smaller organoids at 0.3 x 1000 g for 2 min and discard the medium.
- Dilute the pellet of smaller organoids in 130 µL of 40%-50% basement membrane matrix (diluted with Ad-DF+++ medium12).
- Resuspend well using a 200 µL micropipette.
- Plate the organoids in a 96-well plate
- Using a 20 µL pipette, plate organoids in 32 wells of a pre-warmed 96-well plate. Ensure that each well contains one 4 µL drop of the organoid-matrix solution produced in the previous step.
- The organoids in the solution tend to form a pellet at the bottom of the tube due to gravity-dependent downwards displacement. To prevent this and ensure uniform plating, regularly resuspend the organoid-matrix solution using a 200 µL micropipette (e.g., every time after plating four to eight wells).
- Plate each drop in the center of the well to prevent the drop from running toward the edges of the well, which would reduce image quality later.
- Aim for around 30 organoids per well (minimum 15, maximum 90) without overlapping organoids.
- Keep in mind that these organoids will have to be incubated overnight and will grow slightly during this time, and might start overlapping if the plating density is too high.
- Check plating density using a brightfield microscope with a 5x magnification objective after plating the first one or two wells.
- If the plating density is too high, dilute the organoid-matrix solution stepwise with the addition of the basement membrane matrix until the desired plating density is reached (Figure 2).
- After plating, gently tap the plate on a flat surface to ensure that the majority of organoids are in the same focal plane.
- Incubate the plate
- Incubate the 96-well plate at 37 °C and 5% CO2 during 8-10 min to allow gelation of the basement membrane matrix.
- Add 50 µL of human colon organoid medium +/+12 in each well and incubate overnight for 16-24 h.
- Add no forskolin nor CFTR modulators, so the organoids grow under basal conditions.
3. Organoid imaging using confocal microscopy (day 2)
- Stain the organoids with calcein green
- Prepare a 1 mM stock solution of calcein green by adding 50 µL of dimethylsulfoxide (DMSO) to one vial containing 50 µg of calcein green.
- Prepare a working solution of calcein green by adding 1.2 µL of the stock solution to 200 µL of Ad-DF+++ (concentration 6 µM).
- Add 5 µL of this calcein mixture to each well of the 96-well plate in which organoids were plated (final calcein concentration 0.6 µM). Do not touch and dislocate the matrix drop inside the well when performing this step.
- Rotate and slightly tilt the plate with the lid on a few times to ensure homogenous distribution of the calcein green in the whole well.
- Incubate the plate again for 15-30 min at 37 °C and 5% CO2 to ensure staining of all organoids in the wells.
- Transfer the plate to the confocal microscope
- Transfer the plate to the confocal microscope with an automated stage and integrated incubator. Ensure that the plate is well fixed in the plate holder.
- Incubation at 37 °C and 5% CO2 is optional but not necessary, given the short duration (about 10 min) of the imaging process.
- Focus on the organoids (Figure 3)
- Using the confocal microscope, determine the optimal x/y position and focus (z position) of the organoids in each well manually, and save these positions in the imaging software.
- The best focus implies the sharpest possible delineation of the organoids and visualization of the largest possible part of the organoid drop. If the distribution of the stain is not adequate, repeat steps 3.1.4 and 3.1.5 (incubate, for example, for an extra 5-10 min).
- Use live-cell imaging settings with emission at 488 nm and excitation at 515 nm (specific for visualization of calcein fluorescence) and a 5x LD magnification objective.
- Acquire organoid pictures of the 32 wells with the confocal microscope (Figure 3 and Figure 4).
- Take images in a unidirectional way with a resolution of 1024 pixels x 1024 pixels (pixel size 2.5 µm x 2.5 µm) and depth of 16 bits.
- Choose the laser intensity and master gain for optimal visualization of morphological differences between CF and non-CF organoids (e.g., discrimination between a non-stained water-filled central lumen (if present) and a stained cellular border).
NOTE: Organoids will not be delineated correctly when the master gain and thus the fluorescence signal are too low; setting the master gain too high will result in images with organoids with homogenous very high signal intensity, preventing imaging of more subtle morphological differences (Figure 2). - Save one picture per well for all 32 wells in the microscope format and export them as TIFF files.
4. Image analysis (Figure 5)
- Load the TIFF files in the image analysis software.
- Perform the first quality check based on exclusion criteria determined by the operator (Figure 2): many differentiated or dead structures or debris, inadequate plating density, too many (e.g., overlapping) or too few organoids, and inadequate fluorescence distribution (organoids not clearly delineated, background signal too high).
NOTE: This step can also be performed before exporting images as TIFF files in step 3.4. - Prepare images for analysis
- Recalibrate images, so 1 pixel corresponds to 2.5 µm x 2.5 µm.
- Create and open one Network Data (.ND) file of all 32 pictures for each organoid culture, enabling simultaneous analysis of all 32 pictures per subject.
- Delineate the organoids
- Delineate structures using a lower intensity threshold of 4,500 and an upper threshold of 65,535 (Smooth and Clean functions off; Fill Holes function on; Separate function at x3).
NOTE: This delineates fluorescent structures, with Fill Holes including the lumen in the delineated structure if present.
- Delineate structures using a lower intensity threshold of 4,500 and an upper threshold of 65,535 (Smooth and Clean functions off; Fill Holes function on; Separate function at x3).
- Count the organoids
- Select all structures ≥40 µm.
- Click the Update ND Measurement button (every time a measurement is needed); the counted organoids will be numbered.
NOTE: This equals the total number of organoids in the 32 wells, while small debris, such as dead cells, is excluded.
- Measure intensity and circularity for calculation of the indexes
- Select all structures ≥60 µm and count.
NOTE: This counts the organoids large enough to show morphology typical to either CF or non-CF. Organoids >40 µm and <60 µm are small and dense, both in non-CF and in CF. - Remove all structures touching the borders of the picture. Do this to remove organoids that are not completely visible, as their morphology cannot be accurately quantified.
- Erode 1 pixel (= 2.5 µm) from the border of each ≥60 µm structure. This removes the halo of diffused calcein fluorescence surrounding the organoids.
- Measure the mean intensity of each structure. This way, the mean fluorescence of the organoids is measured.
- Select all structures ≥60 µm again and remove all structures touching the borders.
- Erode 10 pixels (= 25 µm) from the border of each ≥60 µm structure.
NOTE: In non-CF organoids, this erodes the cellular border and leaves only the lumen. In CF organoids, this erodes the outer part of the organoid, which has roughly the same fluorescence as the inner part that remains. - Measure the mean intensity of each eroded structure. This way, the mean fluorescence of the central part of the organoids is measured.
- Measure the circularity of each structure. This corresponds to the mean circularity of the organoids.
- Select all structures ≥60 µm and count.
- Perform the second quality check: exclusion criteria determined by the software. Exclude the set of pictures when less than 50% of organoids (defined as structures ≥40 µm) are ≥60 µm, as enough organoids should be large enough to show morphology typical to either CF or non-CF. Also exclude when <500 or >3,000 organoids (defined as structures ≥40 µm) are present in the 32 wells.
5. Measure the indices in the imaging software (Figure 6)
- Measure the circularity index (CI).
NOTE: This corresponds to the mean circularity measured in step 4.6, which is the mean circularity of all organoids in all 32 wells. CI quantifies the roundness of the organoids, defined as , which is lower in CF than in non-CF organoids
, which is lower in CF than in non-CF organoids - Measure the intensity ratio (IR)
- Calculate the IR by dividing the mean of the intensity measurement after eroding 25 µm from the border of each ≥60 µm structure by the mean of the intensity measurement after eroding 2.5 µm from the border of each ≥60 µm structure.
NOTE: IR measures the presence or absence of a central lumen. IR is equal to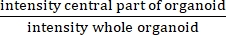 , and is higher in CF than in non-CF organoids.
, and is higher in CF than in non-CF organoids.
- Calculate the IR by dividing the mean of the intensity measurement after eroding 25 µm from the border of each ≥60 µm structure by the mean of the intensity measurement after eroding 2.5 µm from the border of each ≥60 µm structure.
- Simplify the analysis process
- Prepare a standard worksheet using spreadsheet software to automatically calculate these indices upon copying the data (Figure 7).
- To calculate the IR:
- Take the intensity measurement after eroding 2.5 µm from the border of each ≥60 µm structure. Use the mean for the calculation (denominator).
- Take the intensity measurement after eroding 25 µm from the border of each ≥60 µm structure. Use the mean for the calculation (numerator).
- To calculate the CI, take the circularity of all organoids in all wells. The mean corresponds to the CI.
NOTE: When analysis of large batches of images is required, the image analysis process as described in section 4 can be semi-automated, starting from the calibrated ND files and running a macro with all steps combined.
Results
Organoids from 212 subjects were collected during routine clinical visits. No adverse events occurred during or after the rectal biopsy procedure. Organoids were imaged by one researcher blinded to subject characteristics such as genotype and clinical information. Due to low-quality images, 23 subjects were excluded. Examples of successful and failed organoid cultures and image acquisition can be seen in Figure 2.
Organoids of 167 subjects with CF and two disease-...
Discussion
We provide a detailed protocol for rectal organoid morphology analysis (ROMA). The two indexes calculated with ROMA, IR, and CI, distinguished organoids from subjects with CF from those without CF with perfect accuracy. ROMA could thus function as a novel physiological CFTR assay complementary to SCC and other currently available tests13,14,15.
The protocol is dependent on the use of intestinal organo...
Disclosures
This study was funded by the Belgian CF patients' association "Mucovereniging/Association Muco", the Research Grant of the Belgian Society of Paediatrics BVK-SBP 2019, and a grant from the UZ Leuven Fund for Translational Biomedical Research. The authors declare no conflict of interest.
Acknowledgements
We thank the patients and parents who participated in this study. We thank Abida Bibi for all culturing work with the organoids. We thank Els Aertgeerts, Karolien Bruneel, Claire Collard, Liliane Collignon, Monique Delfosse, Anja Delporte, Nathalie Feyaerts, Cécile Lambremont, Lut Nieuwborg, Nathalie Peeters, Ann Raman, Pim Sansen, Hilde Stevens, Marianne Schulte, Els Van Ransbeeck, Christel Van de Brande, Greet Van den Eynde, Marleen Vanderkerken, Inge Van Dijck, Audrey Wagener, Monika Waskiewicz, and Bernard Wenderickx for logistic support. We also thank the Mucovereniging/Association Muco, and specifically Stefan Joris and Dr. Jan Vanleeuwe, for their support and financing. We thank all collaborators from the Belgian Organoid Project: Hedwige Boboli (CHR Citadelle, Liège, Belgium), Linda Boulanger (University Hospitals Leuven, Belgium), Georges Casimir (HUDERF, Brussels, Belgium), Benedicte De Meyere (University Hospital Ghent, Belgium), Elke De Wachter (University Hospital Brussels, Belgium), Danny De Looze (University Hospital Ghent, Belgium), Isabelle Etienne (CHU Erasme, Brussels, Belgium), Laurence Hanssens (HUDERF, Brussels), Christiane Knoop (CHU Erasme, Brussels, Belgium), Monique Lequesne (University Hospital Antwerp, Belgium), Vicky Nowé (GZA St. Vincentius Hospital Antwerp), Dirk Staessen (GZA St. Vincentius Hospital Antwerp), Stephanie Van Biervliet (University Hospital Ghent, Belgium), Eva Van Braeckel (University Hospital Ghent, Belgium), Kim Van Hoorenbeeck (University Hospital Antwerp, Belgium), Eef Vanderhelst (University Hospital Brussels, Belgium), Stijn Verhulst (University Hospital Antwerp, Belgium), Stefanie Vincken (University Hospital Brussels, Belgium).
Materials
| Name | Company | Catalog Number | Comments |
| 1.5 mL microcentrifuge tubes | Sorenson | 17040 | |
| 15 mL conical tubes | VWR | 525-0605 | |
| 24 well plates | Corning | 3526 | |
| 96 well plates | Greiner | 655101 | |
| Brightfield microscope | Zeiss | Axiovert 40C | |
| Centrifuge | Eppendorf | 5702 | |
| CO2 incubator | Binder | CB160 | |
| Computer | Hewlett-Packard | Z240 | |
| Confocal microscope | Zeiss | LSM 800 | |
| Laminar flow hood | Thermo Fisher | 51025413 | |
| Material for organoid culture as detailed in previous protocol10 | |||
| Micropipettes (20, 200, and 1000 µL) | Eppendorf | 3123000039, 3123000055, 3123000063 | |
| Microsoft Excel | Microsoft | Microsoft Excel 2019 MSO 64-bit | Spreadsheet software |
| NIS-Elements Advanced Research Analysis Imaging Software | Nikon | v.5.02.00 | Imaging software |
| Pipette tips (20, 200, and 1000 µL) | Greiner | 774288, 775353, 750288 | |
| Zeiss Zen Blue software | Zeiss | v2.6 | Imaging software |
References
- Riordan, J. R., et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 245 (4922), 1066-1073 (1989).
- Castellani, C., et al. ECFS best practice guidelines: the 2018 revision. Journal of Cystic Fibrosis. 17 (2), 153-178 (2018).
- . CFTR2 Available from: https://www.cftr2.org/ (2022)
- Farrell, P. M., et al. Diagnosis of cystic fibrosis: Consensus guidelines from the cystic fibrosis foundation. The Journal of Pediatrics. 181, 4-15 (2017).
- Vermeulen, F., Lebecque, P., De Boeck, K., Leal, T. Biological variability of the sweat chloride in diagnostic sweat tests: A retrospective analysis. Journal of Cystic Fibrosis: Official Journal of the European Cystic Fibrosis Society. 16 (1), 30-35 (2017).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Boj, S. F., et al. Forskolin-induced swelling in intestinal organoids: An in vitro assay for assessing drug response in cystic fibrosis patients. Journal of Visualized Experiments: JoVE. (120), e55159 (2017).
- Friedmacher, F., Puri, P. Rectal suction biopsy for the diagnosis of Hirschsprung's disease: a systematic review of diagnostic accuracy and complications. Pediatric Surgery International. 31 (9), 821-830 (2015).
- Dekkers, J. F., et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nature Medicine. 19 (7), 939-945 (2013).
- Dekkers, J. F., et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Science Translational Medicine. 8 (344), (2016).
- Vonk, A. M., et al. Protocol for application, standardization and validation of the forskolin-induced swelling assay in cystic fibrosis human colon organoids. STAR Protocols. 1 (1), 100019 (2020).
- Cuyx, S., et al. Rectal organoid morphology analysis (ROMA) as a promising diagnostic tool in cystic fibrosis. Thorax. 76 (11), 1146-1149 (2021).
- Wilschanski, M., et al. Mutations in the cystic fibrosis transmembrane regulator gene and in vivo transepithelial potentials. American Journal of Respiratory and Critical Care Medicine. 174 (7), 787-794 (2006).
- Derichs, N., et al. Intestinal current measurement for diagnostic classification of patients with questionable cystic fibrosis: validation and reference data. Thorax. 65 (7), 594-599 (2010).
- Ramalho, A. S., et al. Correction of CFTR function in intestinal organoids to guide treatment of Cystic Fibrosis. European Respiratory Journal. 57, 1902426 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved