Method Article
Isolation, Expansion, and Nucleofection of Neural Stem Cells from Adult Murine Subventricular Zone
In This Article
Summary
Here, we describe a nucleofection system designed to enhance gene delivery efficiency in expanded neural stem cells (NSCs) isolated from the adult murine subventricular zone. The findings demonstrate that this method significantly improves gene perturbation in NSCs, surpassing the effectiveness of traditional transfection protocols and enhancing cell survival rate.
Abstract
Isolation and expansion of neural stem cells (NSCs) from the subventricular zone (SVZ) of the adult mouse brain can be achieved in a medium supplemented with basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF) as mitogens, producing clonal aggregates known as neurospheres. This in vitro system is a valuable tool for studying NSC potential. Transfection of siRNAs or genes carried in plasmids can be used to induce perturbations to gene expression and study NSC biology. However, the exogenous nucleic acid delivery to NSC cultures is challenging due to the low efficiency of central nervous system (CNS) cells transfection. Here, we present an improved nucleofection system that achieves high efficiency of gene delivery in expanded NSCs from adult murine SVZ. We demonstrate that this relatively simple method enhances gene perturbation in adult NSCs, surpassing traditional transfection protocols with survival rates exceeding 80%. Moreover, this method can also be applied in primary isolated NSCs, providing a crucial advancement in gene function studies through gene expression manipulation via knockdown or overexpression in neurosphere cultures.
Introduction
Neural stem cells (NSCs) are multipotent stem cells resident in the brain. These cells possess the ability to self-renew and differentiate into the three neural lineages: astrocytes, oligodendrocytes, and neurons1. Consequently, NSCs play a crucial role in adult neurogenesis in mammals, a process where new neurons are generated in the brain2. NSCs predominantly reside in two regions within the adult brain termed neurogenic niches: the subventricular zone (SVZ) along the walls of the lateral ventricles and the subgranular zone (SGZ) within the dentate gyrus of the hippocampus3,4. NSCs (also known as B cells) in the SVZ, the most active neurogenic niche in the adult mouse, self-renew and produce transit-amplifying progenitors (TAPs or C cells) that subsequently differentiate into neuroblasts (A cells). These neuroblasts migrate through the rostral migratory stream (RMS) to the olfactory bulbs (OB), where they undergo full differentiation into interneurons, integrating into the pre-existing circuitry3,4,5,6.
Understanding the intricate interplay of molecular cues and signals that regulate NSCs within these niches is important to harness their potential for therapeutic applications. For that purpose, various methods have been developed to study this cell population, ranging from selective primary culture of NSCs to cell selection using surface markers7,8,9,10. The present manuscript details the isolation and culturing of SVZ NSCs in vitro using a serum-free selective medium containing both mitogens: basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF). This medium facilitates cell proliferation and maintains the stemness of NSCs obtained from the SVZ of adult mouse brains forming in vitro clonal, three-dimensional, non-adherent aggregates known as neurospheres9. Neurosphere cultures serve as a controlled platform for manipulating and studying the molecular mechanisms and factors affecting NSC proliferation, self-renewal, differentiation, and survival11. Notably, the number of primary neurospheres formed in the culture allows an estimation of the number of NSCs present in the SVZ in vivo, making it a powerful tool for studying the effects of different conditions on the adult NSC pool12,13. Furthermore, once the primary culture is established, NSCs can generate new aggregates (secondary neurospheres) upon passaging through symmetric divisions under proliferation conditions14. Thus, low-density seeding of secondary neurosphere cells (clonal assay) can be utilized to assess the self-renewal rate of these cultures4,15,16,17.
Despite the potential of neurospheres in uncovering the mechanisms governing NSC regulation, some researchers question the validity of in vitro findings, arguing that the artificial conditions in which cells grow might not faithfully replicate the intricate in vivo microenvironment of neurogenic niches18,19,20,21,22. Another controversial point revolves around the observed heterogeneity in neurospheres. Nonetheless, this variability is believed to mirror the symmetric and asymmetric divisions of the NSCs that naturally occur in vivo23,24. Furthermore, recent validation has supported the utilization of NSC cultures to predict mechanisms operating within the SVZ neurogenic niche in vivo, and several studies have demonstrated that NSCs cultured in vitro accurately maintain the transcriptomic profile observed in vivo11,25.
Therefore, neurosphere cultures not only serve as a method to explore NSC proliferation and differentiation abilities, but also offer a system to study the influence of genes governing NSC biology. A pivotal technique for investigating gene function in NSCs is gene expression perturbation. siRNAs or genes delivered through plasmids can be transfected into cell cultures, resulting in knockdown or upregulation of the target gene. This versatile approach significantly reduces the time and cost compared to establishing cultures using conditional knockout mice, presenting a promising avenue for unraveling the genetic bases of neurogenesis and exploring therapeutic prospects. Altering the expression of specific genes in NSCs enables the modulation of their behavior, influencing crucial biological processes such as proliferation, differentiation, and migration. However, the prospect of transfecting NSCs, particularly within mouse neurospheres, presents notable challenges. The three-dimensional structure of neurospheres compromises the efficiency of transfection, often resulting in low rates of successful exogenous nucleic acid delivery, which limits the extent of genetic manipulation26,27. Additionally, transfection procedures can detrimentally affect cell viability and functionality27. In this context, we present the nucleofection system as a method to mitigate cell damage, achieving a high survival rate and ensuring higher efficacy in gene delivery assays used to perturb NSC cultures.
This manuscript aims to illustrate the procedure for isolating, expanding, and nucleofecting NSCs from the adult SVZ neurogenic niche to perturb genes using the neurosphere culture system. This method surpasses the effectiveness of traditional transfection protocols, presenting significantly higher survival rates and improved gene delivery efficiency among the targeted cells.
Protocol
All experiments performed with animals were previously approved by the University of Valencia's Ethical Committee and authorized by Conselleria de Agricultura, Ganadería y Pesca, Generalitat Valenciana (Spain).
1. Primary culture of neurospheres
- Preparation of reagents
- Prepare a buffer solution of Dulbecco's Phosphate Saline Buffer (DPBS) in deionized water and sterilize by autoclaving. Alternatively, prepare 0.1 M PBS pH 7.4 by adding 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 solution in deionized water.
- Fill two 12-well plates with previously cooled DPBS and keep them on ice: one will be used to preserve the whole brain before dissection, and the other will preserve dissected SVZs until tissue processing.
- Prepare papain-containing enzymatic mix as described below.
- Prepare EDTA/L-cysteine dissociation solution in Earl's Balanced Salt Solution (EBSS) to a final concentration of 0.2 mg/mL each. To achieve complete dissolution, incubate the tube in a water bath at 37 °C.
- Dissolve 12 U/mL of papain in EDTA/L-cysteine solution. Consider that 500 µL of enzymatic solution will be used per brain.
- Introduce papain solution in a water bath 37 °C for around 20 min until the enzyme fully dissolves.
- Filter-sterilize the enzymatic mix with a syringe and a 0.22 µm-pore filter and keep it at 4 °C until use.
NOTE: Papain becomes activated after 30 min at 37 °C, and it can be stored for 12 h at 4 °C.
- Prepare the control culture medium by adding the hormone mix solution according to Table 1 and filter-sterilize it using nitrocellulose 0.22 µm-pore filters. Keep the medium at 4 °C and warm it in a water bath at 37 °C before use.
- Prepare the complete medium by supplementing the control medium with EGF and bFGF just before using as per Table 1. Do not filter the complete medium, as mitogens can be lost with filtering.
- Brain extraction and SVZ dissection
NOTE: Mice are allowed free access to food and water prior to euthanasia. C57BL/6 mice that are 8 to 16 weeks old are used. Both sexes are used without noticeable differences in the preparation.- Sterilize the dissection tools by autoclaving before the surgery. Then, submerge scalpels, tweezers, scissors, and spatulas in a beaker filled with 70% ethanol. Clean every working surface thoroughly with 70% ethanol.
- Sacrifice the mouse by CO2 overdose and confirm by cervical dislocation. Spray the head with 70% ethanol to minimize the possibility of brain tissue contamination.
- Separate the head of the mouse from the rest of the body using scissors. Cut the skin above the skull with small scissors along the rostro-caudal axis until the cranium is completely uncovered.
- Expose the brain by initially cutting the skull along the sagittal suture with small scissors and then, remove the bones using fine tweezers. Be careful not to cause any damage to the brain tissue beneath the skull during this step.
- Carefully extract the brain from the skull using a spatula and place it into a 12-well plate containing cold DPBS. Keep the plate on ice until dissection is completed.
- Transfer the entire brain onto the silicon pad where the dissection will take place and apply a few drops of cold DPBS over the brain (Figure 1A).
- Use a scalpel to remove the OB located at the rostral end of the brain and the cerebellum situated in the caudal part while retaining the central portion of the brain containing both lateral ventricles (Figure 1B).
- Divide the brain along the longitudinal fissure into two hemispheres and then proceed with dissection of both hemispheres separately (Figure 1C).
- Working with one hemisphere, reorient it so that the medial area faces upward. Open the brain along the line of the corpus callosum, separating the cortex and striatum from the hippocampus, septum, and diencephalon, thereby exposing the lateral ventricles (Figure 1D).
NOTE: The SVZ is situated between the ventricle's cavity and the striatum, so the subsequent steps aim to isolate the SVZ as accurately as possible from the surrounding tissue. - Remove the hippocampus, septum, and diencephalon following the ventral limit of the ventricles (Figure 1E,F), the tissue beyond the rostral and caudal ends of the SVZ (Figure 1G), and the cortex following the corpus callosum (Figure 1H,I).
- Tilt the tissue so that the SVZ faces sideways (Figure 1J), allowing the removal of the striatal tissue beneath the SVZ (Figure 1K) to obtain a thin piece of tissue containing the NSCs (Figure 1L).
- Store both dissected SVZs from the same mouse in a 12-well plate containing cold, sterile DPBS until the tissue processing begins.
NOTE: The dissection of both SVZs from one brain should take 10 min. Longer time might affect cell viability. If an increased yield of the cultures is needed, SVZs from multiple mice of the same age, sex, genetic background, and genotype can be pooled.
- Isolation and seeding of NSCs
- Cut each SVZ into 4-5 small pieces to facilitate the disaggregation of the tissue.
NOTE: From this step on, the whole procedure must be performed in sterile conditions in a laminar flow hood. - Transfer the chopped SVZs to a 15 mL centrifuge tube using a sterile plastic Pasteur pipette. Make sure there are no fragments left in the plate.
- Let the tissue sediment at the bottom of the tube and remove the remaining DPBS.
- Add 500 µL of filtered papain-containing enzymatic mix per brain (2 SVZs) and incubate the tubes in a water bath at 37 °C for 30 min.
- After the incubation, add 5 mL of control medium pre-warmed at 37 °C to each sample.
- Centrifuge the samples at 300 x g for 5 min. Then, carefully remove the supernatant using a micropipette or a vacuum pump.
- Add 1 mL of control medium and carefully mechanically disaggregate the pellet, pipetting up and down 10x to 20x, using a fire-polished Pasteur glass pipette until a homogeneous cell suspension is obtained.
CAUTION: This is one of the most critical steps in the protocol, as either excessive or insufficient disaggregation will negatively affect the yield of the cultures. - Add 4 mL of control medium and mix by inversion to wash the cells. Centrifuge at 300 x g for 5 min. Then, remove the supernatant.
- Add 1 mL of complete medium and resuspend the pellet by pipetting up and down 10x to homogenize the cell suspension.
- Count the cell suspension using a Neubauer chamber under the microscope after diluting 1/2 an aliquot of the cell suspension with trypan blue or utilize an automatic cell counter. Make sure that cultures are disaggregated at the single-cell level.
- Once the cell suspension is prepared, seed the obtained cells equally in 8 wells of a 48-well plate in a final volume of 500 µL of complete medium.
NOTE: We usually obtain 20,000 cells from two SVZs, resulting in a final seeding density of approximately 2,500 cells/mL to achieve the maximum yield in cultures.
- Cut each SVZ into 4-5 small pieces to facilitate the disaggregation of the tissue.
- Neurospheres formation
- Incubate cells for 5 to 7 days in vitro (DIV) in a humidified incubator at 37 °C and 5% CO2. Under these conditions, differentiated cells die while NSCs and progenitors proliferate in response to mitogenic stimulation, forming clonal aggregates recognized as primary neurospheres.
NOTE: Either EGF or bFGF can be used to induce neurosphere formation. However, these two growth factors are not entirely substitutable. - Check the size of the neurospheres using an inverted microscope-coupled camera, and once neurospheres reach a size range of 50 to 100 µm, count all primary neurospheres formed in the well at 10x magnification. The total number of primary neurospheres formed serves as an estimate of the number of NSCs present in the SVZ niche.
- After a 10-day period, collect the culture medium from each well to begin passaging. Conduct subsequent passaging after 5 to 7 DIV, although it is worth noting that primary spheres might require a slightly longer duration to reach the ideal size for subculturing (around 100 µm).
- Incubate cells for 5 to 7 days in vitro (DIV) in a humidified incubator at 37 °C and 5% CO2. Under these conditions, differentiated cells die while NSCs and progenitors proliferate in response to mitogenic stimulation, forming clonal aggregates recognized as primary neurospheres.
Table 1: Culture media solutions. The recipe of the control medium is described. A pre-mixed hormone mix solution is made. Stock hormone solutions can be prepared in advance as indicated and stored at -20 °C. Before cell culture, supplement the control medium with EGF and bFGF to prepare complete medium. Storage specifications, stocks, and working concentrations are provided for each component. Please click here to download this Table.
2. Expansion of neurosphere cultures
- Passage of neurospheres
- Collect the medium containing the neurospheres from the multiwell plates and transfer them to a 15 mL centrifuge tube. Ensure the recovery of as many cells as possible by rinsing the wells with a few milliliters of DPBS.
- Centrifuge neurospheres for 5 min at 300 x g. Then, remove the supernatant.
- Add 200 µL of enzymatic solution (see Table of Materials) to the pellet and gently tap the bottom of the tube to dislodge it. Incubate for 10 min at room temperature to facilitate neurospheres dissociation.
- Add 1 mL of control medium to halt the enzymatic reaction and mechanically dissociate neurospheres with a P1000 micropipette by pipetting up and down 10x to 20x.
- Add an additional volume of 4 mL of the control medium and mix by inversion to wash the cells.
- Centrifuge the cells for 5 min at 300 x g. Then, remove the supernatant.
- Add 1 mL of complete medium and resuspend the pellet to homogenize the cellular suspension by pipetting up and down 10x.
NOTE: If the original number and size of neurospheres are too high, add more complete medium to dilute the cell suspension ensuring a reliable cell count. - Count the cell suspension using a Neubauer chamber after diluting 1/2 an aliquot of the cell suspension with trypan blue or utilize an automatic cell counter.
- To expand the cell culture, seed cells at a density of 10,000 cells/cm2 using complete medium in an appropriate culture plate or flask (see Table 2).
- Incubate cells for 5 to 7 DIV in a humidified incubator at 37 °C and 5% CO2 for each passage.
NOTE: Neurosphere cultures can be consistently expanded and propagated through repeated subculturing, not exceeding 10 passages, without altering their proliferation capacities. Either EGF or bFGF can be utilized for culturing; however, long-term expansion of the cultures can only be achieved in the presence of EGF.
- Self-renewal assay
- After passaging neurosphere cultures, plate 1,000 individual cells in a 96-well plate, completing up to a final volume of 200 µL with complete medium, resulting in a final cell density of the cultures of 5 cells/µL.
NOTE: The number of cells seeded must be as accurate as possible. If necessary, prepare intermediate dilutions to ensure a reliable cell suspension volume before plating. Moreover, it is recommended to plate 4 to 5 replicates per condition to reduce technical variability. - Incubate cells at 37 °C and 5% CO2 in a humidified incubator for 5 days or until neurospheres reach an adequate size (60 to 100 µm). Avoid moving or shaking the multiwell plate to prevent neurosphere aggregation.
- After neurosphere formation, count the number of neurospheres formed in each well using an inverted phase-contrast microscope equipped with an attached camera. The self-renewal assay can be conducted using EGF or bFGF separately.
- After passaging neurosphere cultures, plate 1,000 individual cells in a 96-well plate, completing up to a final volume of 200 µL with complete medium, resulting in a final cell density of the cultures of 5 cells/µL.
- Cryopreservation of NSC cultures
NOTE: Neurosphere cultures can be cryopreserved for future use, minimizing the number of mice required. We recommend preserving cells up to passage 10.- To cryopreserve cells, transfer 1 mL of the cell suspension in complete medium to a properly labeled cryotube.
- Add dimethyl sulfoxide (DMSO) to a final concentration of 10% to the cell suspension and gently mix by inverting the tubes.
NOTE: DMSO serves as a cryoprotectant but can be toxic to cells, so this step should be performed swiftly. - Place the tubes in a -80 °C freezer using a freezing container that enables a gradual temperature decrease at 1 °C/min. This gradual freezing process prevents cell damage caused by the formation of ice crystals during cryopreservation. For long-term storage, maintain the tubes in a liquid nitrogen tank at -196 °C.
- Thawing of NSC cultures
NOTE: When required, cells can be thawed, and cultures re-expanded for experiments.- Remove the cryotubes containing frozen NSCs from the liquid nitrogen storage tank and immediately place them in a 37 °C water bath until completely thawed, approximately 5 min.
CAUTION: It is crucial to minimize the bath time since DMSO is toxic and can affect cell viability. - Quickly transfer thawed cells into a 15 mL centrifuge tube containing 5 mL of pre-warmed control medium.
- Centrifuge cells at 300 x g for 5 min and remove the supernatant. Add 1 mL of complete medium and resuspend the pellet by gently pipetting up and down 10x to homogenize the cellular suspension before seeding.
NOTE: Thawing can affect cell viability; therefore, it is recommended to passage cells at least once before seeding for new experiments.
- Remove the cryotubes containing frozen NSCs from the liquid nitrogen storage tank and immediately place them in a 37 °C water bath until completely thawed, approximately 5 min.
- Mycoplasma testing
NOTE: Mycoplasma contamination can compromise cell health, affecting growth rates. Therefore, mycoplasma testing is crucial to ensure the integrity and reliability of experiments.- DNA extraction
- Collect a minimum of 100 µL of the cell culture for DNA extraction. Both supernatant and cell pellet can be used for DNA extraction. Store this aliquot at -20 °C until extraction.
- Pierce the tube lid and boil the aliquot at 99 °C for 15 min using a thermoblock.
NOTE: Piercing of tube lids avoids tubes opening during boiling at high temperatures. - Centrifuge for 5 min at 16,900 x g. Transfer the supernatant containing the DNA to a new tube. Store the tube with the DNA at -20 °C until PCR testing.
- PCR for Mycoplasma detection
- Prepare the PCR mix using the following components: 8.9 µL of nuclease-free water, 3 µL of 5x reaction buffer, 1.2 µL of 25 mM MgCl2, 0.25 µL of 2.5 mM dNTPs, 0.15 µL of Taq polymerase and 0.25 µL of each mycoplasma-specific primer at a concentration of 10 µM (Forward, Fw: 5'GATGTTTAGCCGGGTCGAGAG3' and Reverse, Rv: 5'GATGTCAAGAGTGGGTAAGGTT3').
- Add 1 µL of genomic extracted DNA. Prepare the PCR mix in a laminar flow hood to prevent DNA contamination.
- Incubate the reaction in a thermal cycler as follows: initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 53 °C for 1 min, extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min.
- To identify the size of the resulting band (around 500 bp), load 15 µL of the DNA PCR product mixed with 3 µL of 6x loading dye solution onto a 2% agarose gel and run the electrophoresis for 40 to 50 min at 100 V.
- DNA extraction
Table 2: Plates and flasks used for culturing. The dimensions and volumes of the most commonly used seeding plates and flasks are provided. The table includes the diameter, growth area, and medium volume used for each vessel, along with an example of the number of NSCs to be seeded under expansion conditions (10,000 cells/cm2). Please click here to download this Table.
3. Nucleofection of neurosphere cultures
- Preparation of reagents
- DNA preparation
- Prepare 25 µL of competent E. coli DH5α cells in a 1.5 mL tube for transformation.
- Add 1 µL of the plasmid of interest (at 100 to 200 ng/µL) to the cells and subject them to a thermal shock at 42 °C for 45 s. Here, a plasmid containing a short hairpin (sh)RNA for silencing Snrpn expression and a control plasmid shSCRAMBLE are used.
NOTE: Keep competent cells on ice throughout the procedure to maintain transformation effectiveness. - Plate the transformed cells on LB agar plates with the appropriate antibiotic and incubate them at 37 °C for 18 h. Keep the Petri dishes inverted to prevent droplet formation due to condensation, which might disturb bacterial colonies.
NOTE: Add appropriate antibiotics to the LB culture medium to exclusively allow the growth of transformed bacteria. - Using a sterile pipette tip, pick individual bacterial colonies and transfer them to an Erlenmeyer flask previously filled with 250 mL of LB to initiate a liquid culture. Avoid picking small satellite colonies positioned around the larger target colonies, as they might not have incorporated the desired plasmid.
- Incubate the flask while vigorously shaking on an orbital shaker at 37 °C overnight.
- Utilize an endotoxin-free maxiprep kit and extract pure plasmid DNA from the bacterial cultures following the manufacturer's instructions. Reconstitute the DNA in the appropriate volume of endotoxin-free water to obtain a plasmid concentration of 1 to 2 µg/µL.
NOTE: The maxiprep yield depends on the culture growth. Usually, resuspending the plasmid DNA pellet in a volume of 200 µL will result in the desired DNA concentration. - Store the tube with the plasmid DNA at -20 °C until use. Prepare a 1.5 mL tube with a final volume of 5 µL containing 2 to 6 µg of the plasmid DNA intended for nucleofection.
- Material preparation
- Prepare the nucleofection solution. If using the recommended kit (see Table of Materials), prepare 95 µL of the nucleofection solution in a 1.5 mL tube for each desired nucleofection. Pipettes and cuvettes are usually included in the kit. Prepare one of each for every nucleofection condition.
- Prepare a T25 flask filled with 4 mL of pre-warmed complete medium per condition and keep it in the incubator until use.
- DNA preparation
- Nucleofection of NSCs
- Use 1 to 2 x 106 individual cells per condition for assessment. Centrifuge the cells for 5 min at 300 x g, then remove the supernatant.
- Add 1 mL of DPBS and resuspend the pellet by pipetting up and down 10x to ensure a homogeneous cellular suspension.
- Repeat centrifugation (step 2.1.6) for a second wash and finally resuspend the pellet in 95 µL of nucleofection solution.
NOTE: Washing steps are crucial to remove antibiotics from the control medium and ensure effective nucleofection. - Combine the 95 µL of cells with a premixed solution containing 5 µL of each plasmid (2 µg total) selected for nucleofection.
- Using a P200 micropipette, transfer this solution to a cuvette and place it in the nucleofector device.
- Nucleofect the cells with the desired plasmids using the optimized NSC program of the nucleofector system.
- After completing the electroporation, carefully introduce warm complete medium to the cuvette using the Pasteur pipettes provided in the kit. Subsequently, transfer the contents of the cuvette to a previously prepared T25 flask containing pre-warmed complete medium. After electroporation, there might be cell death, DNA precipitation, and the formation of a viscous aggregate. Avoid transferring this to the T25 flask, as it might reduce cell survival.
NOTE: This step is crucial and should be performed as quickly as possible to ensure a high yield of nucleofection and subsequent culture survival. - Incubate nucleofected cells at 37 °C and 5% CO2 in a humidified incubator for 3 to 5 days.
- Selection of nucleofected cells
- Strategy A: Flow cytometry selection by fluorescence-activated cell sorting (FACS)
- After 3 to 5 days of nucleofection, passage the nucleofected cultures following step 2.1 and collect all individualized cells obtained.
- Filter samples through a 40 µm cell strainer to remove cell aggregates and analyze them with the cell sorter flow cytometer.
NOTE: It is crucial to sort the same number of cells in each condition to obtain comparative results. - Using the cytometer's software, gate alive cells based on two measures: forward scattered light (FSC-A) that corresponds to cell size and side scattered light (SSC-A) that corresponds to intracellular complexity. Select the population of living cells characterized by high FSC-A and high SSC-A.
NOTE: Optionally, add 0.1 µg/mL of 4′,6-diamidino-2-phenylindole (DAPI) to cell suspension before sorting. This will allow the discarding of dead cells and apoptotic bodies from the gating. - Discard cell doublets and triplets by plotting FSC-A versus FSC-H parameters. Choose the events included in the diagonal of the plot, representing singlets.
NOTE: When cells pass through the laser, the cytometer detects voltage changes over time. FSC-A represents the signal area, and FSC-H signifies peak height. Singlets show a proportional relationship between FSC-A and FSC-H (located on the diagonal of the plot), while cell aggregates display a greater FSC-A relative to FSC-H due to a longer signal. - Select nucleofected cells based on the fluorescence intensity of the reporter gene encoded by the plasmid used. For instance, when using GFP+ cells can be selected based on FITC-A fluorescence levels versus SSC-A plot.
- Isolate the desired number of target cells for each condition and collect them directly in a collection tube filled with complete medium.
- Seed nucleofected NSCs at a density of 10,000 cells/cm2. Incubate the sorted cell cultures at 37 °C and 5% CO2 in a humidified incubator.
- Strategy B: Selection using antibiotic resistance encoded in the plasmid
- After 48 h of nucleofection, add the appropriate antibiotic concentration to select resistant cells.
NOTE: Before starting, it is recommended to test multiple concentrations of the antibiotic on non-nucleofected cells. The optimal antibiotic concentration is the lowest amount at which all non-resistant cells are eliminated. For example, we determined an optimal concentration of 4 µg/mL antibiotic blasticidin for NSC cultures. - Incubate the cells at 37 °C and 5% CO2 in a humidified incubator for 48 h.
- After 48 h of antibiotic treatment, subculture the nucleofected cells according to step 2.1. If the neurospheres remain small, remove the antibiotic from the culture medium by centrifugation at 300 x g for 5 min. Resuspend the cells and culture them in a flask using only complete medium. Incubate the cells at 37 °C and 5% CO2 in a humidified incubator until they can be expanded following step 2.1.
- After 48 h of nucleofection, add the appropriate antibiotic concentration to select resistant cells.
- Strategy A: Flow cytometry selection by fluorescence-activated cell sorting (FACS)
Results
Optimal culture conditions enable isolation and expansion of adult SVZ-derived NSCs in vitro
Cultures of NSCs derived from the adult SVZ have served as a valuable in vitro method for investigating the molecular mechanisms and niche signals regulating NSCs within their specific microenvironments. The neurosphere assay outlined in this manuscript was employed to examine the NSC count within the adult SVZ. SVZ tissue was isolated from the brains of 3-month-old mice, dissociated, and cultured in complete NSC medium supplemented with both EGF and bFGF or each separately. After 10 days in vitro (DIV), the total count of primary spheres formed under these three distinct culture conditions was quantified using phase contrast microscopy (Figure 2A,B). Remarkably, our findings demonstrate that the presence of EGF led to maximal primary sphere formation, which is evident from the reduced count of primary spheres observed in cultures with only bFGF (Figure 2A,B). To evaluate the self-renewal capacity of NSCs under varied medium conditions, cells were subcultured and plated at low density (5 cells/µL) in media supplemented with the aforementioned combinations of mitogens. Secondary spheres quantification revealed that SVZ NSCs need at least EGF to efficiently self-renew and that the combination of both EGF and bFGF ameliorates the self-renewing capacity of cells (Figure 2B). Moreover, for a detailed analysis of growth dynamics across diverse media conditions, the number of cells seeded and obtained throughout 7 passages was recorded. Growth curves obtained from different media conditions confirmed that cultures supplemented with EGF alone or in combination with bFGF exhibited improved growth dynamics compared to cultures solely supplemented with bFGF (Figure 2C). Collectively, these findings substantiate that concurrent use of both EGF and bFGF enhances NSC culturing yield in vitro.
In order to investigate the NSC count within the SVZ across different postnatal ages and evaluate the impact of mice aging on neurosphere culture efficiency, SVZ tissue from mice aged 1 month up to 12 months was dissected. Notably, our findings revealed a significant decline in the number of primary spheres with increasing age, showcasing maximum efficiency in sphere count at around 2 to 4 months of age (Figure 2D). Additionally, to evaluate the self-renewal capacity of NSCs during subculturing, NSCs from mice aged 2 to 4 months were subcultured and seeded at clonal density (5 cells/µL) in complete medium. Quantification of the number of secondary spheres throughout subsequent passages indicated a considerable decrease in culture efficiency over passages (Figure 2E). Based on all these observations, conducting self-renewal assays during early passages is recommended to further optimize NSC culture conditions.
Nucleofection is a highly efficient technique for manipulating gene expression in adult NSCs
Given that NSCs are not easily transfectable, to manipulate gene expression, here we present a protocol of nucleofection with a higher rate of successful gene delivery without the need to use viral transduction. Following dissection and culturing of individual SVZs from 2 to 4-month-old mice, NSCs were nucleofected with a GFP-carrying plasmid as described (Figure 3A). Detection of GFP-positive cells 2 days post-nucleofection revealed an efficiency ranging from 30% to 50%, consistent with previous literature28. To specifically isolate successfully nucleofected NSCs, cells were sorted by FACS based on GFP fluorescence intensity 3 to 5 days post-nucleofection (Figure 3A-C). Approximately 40% of pre-FACS analyzed cells exhibited high GFP fluorescence levels and were subsequently selected by sorting (Figure 3C). Reseeding of sorted GFP+ NSCs showed that all cells were GFP positive, validating the nucleofection-based cell isolation method (Figure 3D). Notably, nucleofected NSCs maintained viability through subsequent passages, confirming nucleofection feasibility for adult NSCs. These results emphasize the effectiveness of combining nucleofection and FACS to establish a pure culture of modified NSCs.
As an example of gene modulation in adult NSCs, gene attenuation via short hairpin (sh) RNA nucleofection was employed. Specifically, a shRNA targeting the Small nuclear ribonucleoprotein polypeptide N (Snrpn) gene, carrying a CAG-GFP reporter (shSNRPN) was nucleofected in NSC cultures derived from 2-month-old mice. Downregulation of Snrpn was confirmed by qPCR and immunocytochemistry in cells nucleofected with the shSNRPN plasmid but not for the control shSCRAMBLE plasmid (Figure 3E,F)29. To elucidate the effects of Snrpn downregulation on NSC self-renewal capacity, a low-density assay was performed in nucleofected cells (Figure 3F,G). Quantification of secondary neurospheres in nucleofected cultures revealed an increased neurosphere formation capacity upon Snrpn downregulation (Figure 3G). This assay underscores the nucleofection capacity to manipulate gene expression in adult NSCs and, more specifically, identifies the role of Snrpn in maintaining stemness in adult NSCs.
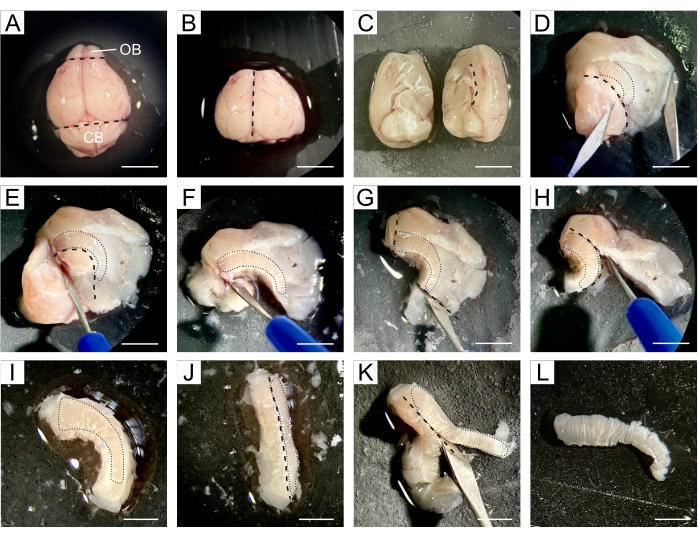
Figure 1: Detailed description of SVZ dissection. (A) After removing the mouse brain, the entire brain is transferred with DPBS to a silicon pad for dissection. (B) OB and cerebellum are removed from the brain using a scalpel. (C) The brain is divided into two hemispheres to proceed with dissection separately. (D) The brain is opened along the line of the corpus callosum, separating the cortex and striatum from the hippocampus, septum, and diencephalon, thereby exposing the lateral ventricles. (E-F) The hippocampus, septum, and diencephalon are removed following the ventral limit of the ventricles. (G-I) The tissue beyond the rostral and caudal ends of the SVZ and the cortex is removed following the corpus callosum. (J-K) The tissue is tilted so that the SVZ faces sideways, allowing the removal of the striatal tissue beneath the SVZ. (L) A thin piece of tissue containing the SVZ is obtained. Black dashed lines indicate the cutting site. Black dotted lines indicate the location of the SVZ at each step. Abbreviations: OB = olfactory bulb; CB = cerebellum. Scale bar in A-H: 5 mm; in I-L: 3 mm. Please click here to view a larger version of this figure.
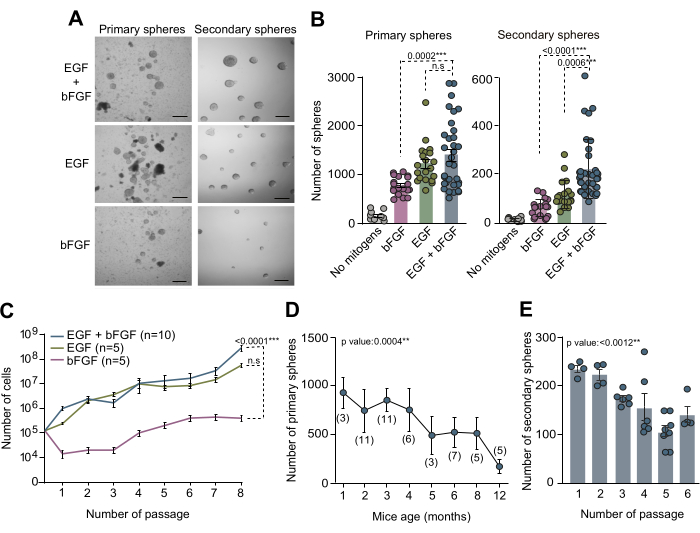
Figure 2: Both mitogens EGF and bFGF are necessary for an optimal culture and expansion of NSCs in vitro. (A) Phase-contrast microscopy images of primary and secondary neurosphere cultures in NSC medium supplemented with both EGF and bFGF or each mitogen separately. (B) Number of primary spheres obtained per mouse and per well based on the mitogen composition of the culture medium: no mitogens (gray, n=9), EGF+bFGF (blue, n=33), only EGF (green, n=20), or only bFGF (pink, n=19; left panel). Number of secondary neurospheres formed at low density (5 cells/μL) per well based on the mitogen composition of the culture medium): no mitogens (gray, n=11), bFGF (pink, n=18), EGF (green, n=18) or EGF+bFGF (blue, n=37; right panel). The Kruskal-Wallis test with Dunn's post hoc analysis was used. (C) Growth curves showing the total number of cells formed after 8 passages in distinct neurosphere cultures according to the mitogens present in the culture medium: bFGF (pink, n=5), EGF (green, n=5) or EGF+bFGF (blue, n=10). Linear regression analysis was used. (D) The number of primary neurospheres derived from individual mice dissected and disaggregated from animals at various postnatal development stages. Linear regression analysis was used. n indicated in brackets. (E) The number of secondary neurospheres formed at low density (5 cells/µL) through subsequent passages in vitro to assess their self-renewal capacity. The Kruskal-Wallis test was used, and p values are included: *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. n.s: non-significant. Error bars represent the SEM. Scale bar in A is 100 µm. Please click here to view a larger version of this figure.
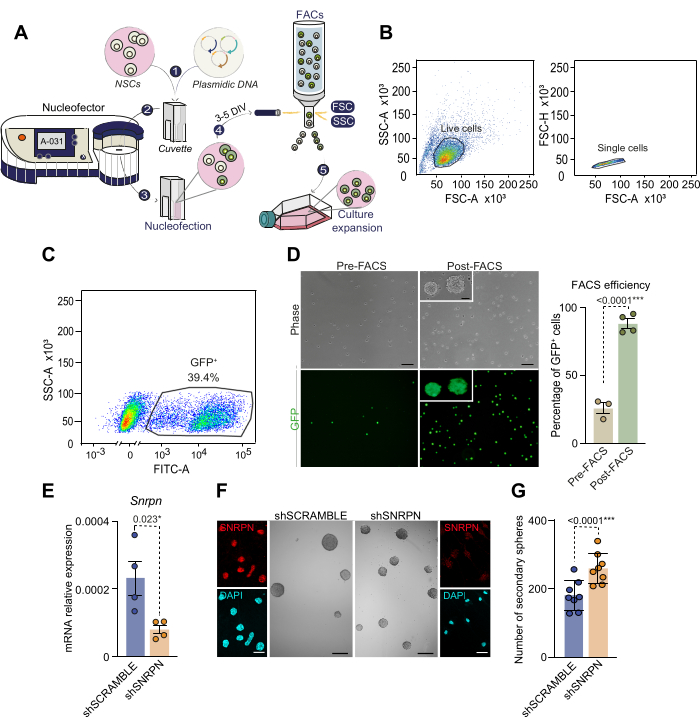
Figure 3: Schematic of the nucleofection process and FACS-based cell selection of nucleofected cells. (A) Overview of the nucleofection process and FACS-based cell selection of nucleofected cells. 1) Plasmid DNA solution is combined with the nucleofection solution and added to the cells. This mixture is then transferred to a cuvette for nucleofection. 2) The cuvette containing DNA and cells is inserted in the nucleofector, where an electric shock is applied. For neurosphere cultures isolated from the adult SVZ, we recommend using the A-031 NSC program. 3) Following nucleofection, cells are transferred to a flask containing complete medium supplemented with EGF and bFGF. 4) Cells are sorted by FACS based on the expression of the reporter contained in the plasmid DNA. 5) Sorted cells undergo further culture for subsequent experiments.(B) FACS-based selection strategy and gating for nucleofected NSCs. First, living cells are selected based on high FSC-A and high SSC-A (left graph), and then cell doublets and triplets are eliminated by analyzing FSC-A versus FSC-H parameters (right graph). (C) FACS gating for nucleofected cells based on their GFP expression (SSC-A versus FITC-A fluorescence). (D) Phase-contrast microscopy and GFP fluorescence images of nucleofected NSCs before (n=3) and after (n=4) FACS selection (left panel). The percentage of nucleofected GFP+ cells is depicted before and after FACS selection (right panel). t-test was used. (E) Quantitative PCR (qPCR) analysis of Snrpn expression in proliferating NSCs nucleofected with Snrpn-specific shRNA (n=4). Nucleofection using a shRNA SCRAMBLE served as the control (n=4). t-test was used. (F) Immunocytochemistry images displaying SNRPN (in red) in NSCs 7 days after the shRNA nucleofection (outer panels). DAPI was utilized to counterstain DNA. Representative images depicting neurospheres derived from NSCs under shSNRPN and shSCRAMBLE conditions (middle panels). (G) Quantification of the number of neurospheres formed in NSC cultures after nucleofection with shSNRPN (n=8) or shSCRAMBLE (n=8). t-test was used. p values are included: *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. n.s: non-significant. Error bars represent the SEM. Scale bar in D, 10 µm; in F, 100 µm (high magnification images in F, 7 µm). Please click here to view a larger version of this figure.
Discussion
Due to the lack of definitive markers for identifying the NSC population in vivo, the analysis of NSCs has been primarily based on observing the behavior of cells isolated from neurogenic niches in ex vivo conditions. Pioneering work by Reynolds and Weiss laid the groundwork by establishing precise culture conditions, enabling the isolation and expansion of individual cells from young adult (2 months old) mouse SVZ tissue under non-adhesive conditions. These cells are usually propagated in a serum-free medium containing EGF and bFGF, conditions that entirely prevent differentiation into neurons and glia while promoting proliferation. Indeed, under these culture conditions9, most cells die during the first days in culture, but a small subset begins dividing and primarily forms floating neurospheres9. Enzymatic dissociation and subculture of these cell aggregates facilitate culture propagation, demonstrating the self-renewal capacity of these cultures.
Notably, neurosphere cultures show expansion potential with the addition of EGF only, while NSCs grown in a medium containing only bFGF do not show long-term cell proliferation30. Both pieces of evidence point out EGF as the primary NSC self-renewal mitogen. Nevertheless, the presence of both EGF and bFGF in the culture medium improves the self-renewal capacity of NSCs31, as well as contributes to balancing the differentiation potential into astrocytes, neurons, and oligodendrocytes32,33,34,35. Moreover, the use of a defined mixture of hormones and factors instead of commercial alternatives to supplement the medium ensures the high quality and reproducibility of NSC cultures. Under these conditions, mouse NSC cultures can persist as stable cell lines without undergoing immortalization. Nevertheless, a concern in neurosphere cultures is the potential of proliferative cell populations to undergo genetic transformations, bypassing cell cycle regulatory mechanisms and leading to an immortalized phenotype. Therefore, although neurosphere cultures are highly expandable, cultures beyond 10 in vitro passages are usually discarded for studies, as they may undergo replicative aging, and cells with abnormal chromosomal content or irregular growth dynamics might emerge. Moreover, the use of long-term established neurosphere cultures must be continuously monitored. Neurosphere cultures also offer the opportunity to examine their characteristics and potential in a controlled environment, providing a more precise and adjustable setting that can be modulated and monitored more accurately than in vivo. Through clonogenic or population analyses in vitro, it is possible to quantify the self-renewal and proliferation capacities of these cells, facilitating the identification of the underlying mechanisms governing these properties.
Nevertheless, despite the large list of advantages of working with NSCs in vitro, the nature of this culturing protocol and the delicacy of NSCs constitute a challenge in the field. For example, the number of primary neurospheres generated during a typical dissection can vary significantly based on the skill and precision of the experimenter. The primary neurosphere results illustrate this variability in the number of primary neurospheres obtained from the SVZ of 2-month-old mice, ranging between 500 and 3000 neurospheres. Various factors may contribute to this variability. First, the precision of the dissection minimizes unwanted parenchymal tissue, which inhibits primary sphere formation. Second, generating small pieces of SVZ tissue allows for efficient enzymatic digestion and trituration, thereby reducing cell loss. This highlights the need for sufficient prior practice of the dissection protocol and a fine-tuned development of tissue processing during the establishment of the neurosphere cultures.
Another limitation of these cultures lies in the fact that neurospheres can comprise both NSCs and progenitor cells, making it challenging to distinguish between these two populations within primary cultures. While various markers like GFAP, Nestin, Musashi, and SOX2 have been reported to be expressed by NSCs8, none of these have been exclusively associated with NSCs. Emerging FACS techniques based on cell surface antigen expression have enabled the isolation of NSCs and their progeny. These studies have demonstrated that transit-amplifying progenitors are unable to form neurospheres upon passage25,36. Thus, while the relationship between SVZ cells and neurosphere-initiating cells requires further refinement18,19,20,21,22,23, the ability of neurospheres to be serially passaged over an extended period may reflect the presence of NSCs in the cultures.
This neurosphere culture system has served as a robust model for investigating the impact of signaling pathways and gene expression in maintaining NSC self-renewal capacity in vitro15,29. One approach to explore these aspects involves transfecting NSCs to either overexpress or knock down specific genes. This can be accomplished through various techniques, including viral and non-viral methods. While viral vectors often achieve high gene transfer efficiency, they have important limitations, such as the high safety requirements and time-consuming vector production26. Conversely, classical transfection methods like lipofection and electroporation achieve very low transfection rates, making them unfeasible for hard-to-transfect cells. The nucleofection technology offers a user-friendly approach by combining cell type-specific nucleofection solutions with unique electrical parameters for each cell type37,38. This ensures DNA transfer directly into the cell nucleus39, allowing the DNA incorporation in a cell cycle-independent manner. Consequently, nucleofection emerges as a viable technique for transfecting difficult-to-transfect cells such as mouse NSCs28,40.
One limitation of this method is that a minimum of 2 x 106 cells is recommended and necessary for each nucleofection, although in optimal conditions, a lower number of cells can be used per single nucleofection (i.e., 5 × 105 cells). Another drawback of the method is that the quality and concentration of DNA used for nucleofection significantly influence gene transfer efficiency. Utilizing endotoxin-free prepared DNA is highly recommended to prevent elevated cell mortality due to endotoxin presence. Additionally, using more than 6 µg of total DNA for nucleofection can substantially reduce both gene transfer efficiency and cell viability. Finally, electric pulse administration is also critical for the survival of nucleofected cells.
In this work, we exemplified the manipulation of Snrpn gene expression by employing a strategy involving the downregulation of its expression using episomal plasmids. These plasmids are not integrated into the genome of the cells, and as NSCs keep proliferating in vitro, the introduced DNA will be subsequently diluted along cell division and, thus, lose the effect of genetic manipulation. Therefore, this strategy is valuable to study the effect of an acute alteration in a short period or to birthdate cells in vitro. Some alternatives to evaluate a more prolonged effect of gene perturbation are available. For example, an integrative system such as the transposon-based piggyBAC could be used. This system consists of introducing the desired coding sequence contained in the plasmid flanked by transposable sequences and co-nucleofect the cells with a plasmid containing the sequence for the enzyme transposase41. Alternatively, transposons or the CRISPR/Cas systems could be used.
Further development of transfecting technologies will be an important step towards high throughput assays to assess the role of different genes on adult neural stem cell physiology. In combination with increasingly sophisticated purification and expansion methods, these studies will enable the understanding of the in vitro biology of adult NSCs and the comparison of the biological differences between floating neurospheres and NSCs in vivo.
Disclosures
The authors declare no competing financial interests.
Acknowledgements
This work was supported by grants from Ministerio de Ciencia e Innovación and Agencia Estatal de Investigación (MCIN/AEI) (PID2019-110045GB-I00; PID2022-142734OB-I00 and EUR2023-143479) to SRF. EJV (FPU20/00795) and DSL (FPU22/03797) are funded by the Spanish Formación de Profesorado Universitario fellowship program (FPU). LLC (PRE2020-094137) and JDM (PREP2022-000680) are funded by the Spanish Formación de Personal Investigador (FPI) fellowship program. CMM (CIACIF/2022/366) is funded by Generalitat Valenciana. MIL (CPI-22-481) is funded by Programa INVESTIGO fellowship (Next Generation EU). Open Access funding provided by the Ministerio de Ciencia e Innovación.
Materials
| Name | Company | Catalog Number | Comments |
| 0.22 μm pore-filter bottles (250 mL) | VWR | 514-0330P | |
| 0.22 μm pore-filter bottles (500 mL) | VWR | 514-0332P | |
| 12-well plate | LabClinics | PLC30012 | |
| 15 mL tube | Fisher | 10738771 | |
| 24-well plate | LabClinics | PLC30024 | |
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) | BioWest | L0180-100 | |
| 4′,6-diamidino-2-phenylindole (DAPI) | Sigma | D9542 | |
| 48-well plate | ThermoFisher | 150687 | |
| 6-well plate | LabClinics | PLC30006 | |
| 96-well plate | Labclinics | PLC30096 | |
| Accutase solution | Sigma | A6964-100ML | Referred as enzymatic solution |
| Amaxa Mouse Neural Stem Cell Nucleofector Kit | Lonza | VPG-1004 | |
| Amaxa Nucleofector Iib | Lonza | 10700807 | |
| Antibiotic/Antimitotic (A/A) | Sigma | A5955 | |
| Apo-t-Transferrine | Sigma | T2252-1G | |
| Basic Fibroblast Growth Factor (bFGF) | Sigma | F0291 | |
| Blasticidin | Sigma | 203350 | |
| Bovine Serum Albumin (BSA) | Sigma | B4287-25MG | |
| Cell strainer 40 μm | LabClinics | PLC93040 | |
| Deoxynucleotide triphosphate (dNTPs) | NZYTech | MB08701 | |
| Dimethyl sulfoxide (DMSO) | Sigma | D8418 | |
| Dulbecco’s Phosphate Saline Buffer (DPBS) | Gibco | 14080-055 | |
| Dulbecco's Modified Eagle's Medium (DMEM) F12 1:1 | Gibco | 11320-074 | |
| E. coli DH5α Competent Cells | ThermoFisher | EC0112 | |
| Earles's Balanced Salt Solution (EBSS) | Gibco | 24010-043 | |
| Epidermal Growth Factor - Human recombinant (EGF) | Gibco | 53003-018 | |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma | E-6511 | |
| Fine Forceps | Fine Science Tools | 11274-20 | |
| Fine Scissors Sharp | Fine Science Tools | 14060-09 | |
| Glucose | Sigma | G-7021 | |
| GoTaq G2 Flexi DNA Polymerase | Promega | M7805 | Kit includes reagents for PCR |
| Heparin | Sigma | H-3149 | |
| Insuline | Sigma | I6634 | |
| LB agar (Lennox) | LabKem | AGLB-00P-500 | |
| LB broth (Lennox) | LabKem | LBBR-00P-500 | |
| L-Cysteine | Sigma | C-8277 | |
| L-Glutamine | Gibco | 25030-024 | |
| Neubauer chamber | Blaubrand | BR718620 | |
| Nuclease free water | Labbox | WATR-00A-10K | |
| NZYMaxiprep Endotoxin Free Kit | NZYTech | MB39901 | |
| Papain Lyophilized | Worthington | LS003119 | |
| Progesterone | Sigma | P-6149 | |
| Putrescine | Sigma | P-7505 | |
| Scalpel | Fine Science Tools | 10316-14 | |
| shSCRAMBLE | Mission (Sigma) | SHC003 | |
| shSNRPN | Mission (Sigma) | TRCN0000109285 | |
| Sodium Selenite | Sigma | S-9133 | |
| Spatula | Fine Science Tools | 10090-17 | |
| Sterile PES Syringe Filters (0.22 μm pore-filter) | Epica | SFPE-22E-050 | |
| T12.5 cm2 Flask | Biofil | TCF012025 | |
| T25 cm2 Flask | LabClinics | PLC70025 | |
| T75 cm2 Flask | LabClinics | PLC70075 | |
| Tweezers | Fine Science Tools | 91150-20 |
References
- Gage, F. H. Mammalian neural stem cells. Science. 287 (5457), 1433-1438 (2000).
- Ming, G., Song, H. Adult neurogenesis in the mammalian central nervous system. Ann Rev Neurosci. 28 (1), 223-250 (2005).
- Doetsch, F., Caillé, I., Lim, D. A., García-Verdugo, J. M., Alvarez-Buylla, A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 97 (6), 703-716 (1999).
- Obernier, K., et al. Adult neurogenesis is sustained by symmetric self-renewal and differentiation. Cell Stem Cell. 22 (2), 221-234 (2018).
- Doetsch, F., García-Verdugo, J. M., Alvarez-Buylla, A. Regeneration of a germinal layer in the adult mammalian brain. Proc Natl Acad Sci. 96 (20), 11619-11624 (1999).
- Doetsch, F., García-Verdugo, J. M., Alvarez-Buylla, A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 17 (13), 5046-5061 (1997).
- Mamber, C., Kozareva, D. A., Kamphuis, W., Hol, E. M. Shades of gray: The delineation of marker expression within the adult rodent subventricular zone. Prog Neurobiol. 111, 1-16 (2013).
- Cebrian-Silla, A., et al. Single-cell analysis of the ventricular-subventricular zone reveals signatures of dorsal and ventral adult neurogenesis. eLife. 10, e67436(2021).
- Reynolds, B. A., Weiss, S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 255 (5052), New York, N.Y. 1707-1710 (1992).
- Azari, H., Rahman, M., Sharififar, S., Reynolds, B. A. Isolation and expansion of the adult mouse neural stem cells using the neurosphere assay. J Vis Exp. (45), e2393(2010).
- Jensen, J. B., Parmar, M. Strengths and limitations of the neurosphere culture system. Mol Neurobiol. 34 (3), 153-161 (2006).
- Ferrón, S. R., et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 475 (7356), 381-385 (2011).
- Gil-Perotín, S., et al. Adult neural stem cells from the subventricular zone: A review of the neurosphere assay. Anat Record. 296 (9), 1435-1452 (2013).
- Obernier, K., Alvarez-Buylla, A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 146 (4), 156059(2019).
- Ferrón, S. R., et al. A combined ex/in vivo assay to detect effects of exogenously added factors in neural stem cells. Nat Prot. 2 (4), 849-859 (2007).
- Ferron, S., et al. Telomere shortening and chromosomal instability abrogates proliferation of adult but not embryonic neural stem cells. Development. 131 (16), 4059-4070 (2004).
- Singec, I., et al. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat Meth. 3 (10), 801-806 (2006).
- Santa-Olalla, J., Baizabal, J., Fregoso, M., Cárdenas, M. D. C., Covarrubias, L. The in vivo positional identity gene expression code is not preserved in neural stem cells grown in culture. Eur J Neurosci. 18 (5), 1073-1084 (2003).
- Pastrana, E., Silva-Vargas, V., Doetsch, F. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 8 (5), 486-498 (2011).
- Wan, F., et al. The utility and limitations of neurosphere assay, CD133 immunophenotyping and side population assay in Glioma stem cell research. Brain Pathol. 20 (5), 877-889 (2010).
- Deleyrolle, L. P., Rietze, R. L., Reynolds, B. A. The neurosphere assay, a method under scrutiny. Acta Neuropsych. 20 (1), 2-8 (2008).
- Campos, L. S. Neurospheres: Insights into neural stem cell biology. Neurosci Res. 78 (6), 761-769 (2004).
- Reynolds, B. A., Rietze, R. L. Neural stem cells and neurospheres—re-evaluating the relationship. Nat Meth. 2 (5), 333-336 (2005).
- Parker, M. A. Expression profile of an operationally-defined neural stem cell clone. Exp Neurol. 194 (2), 320-332 (2005).
- Belenguer, G., et al. Adult neural stem cells are alerted by systemic inflammation through TNF-α receptor signaling. Cell Stem Cell. 28 (2), 285-299 (2021).
- Cruz, M. T. G., Simões, S., Lima, M. C. P. de Improving lipoplex-mediated gene transfer into C6 glioma cells and primary neurons. Exp Neurol. 187 (1), 65-75 (2004).
- Hagemann, C., et al. High efficiency transfection of glioma cell lines and primary cells for overexpression and RNAi experiments. J Neurosci Meth. 156 (12), 194-202 (2006).
- Moritz, S., Lehmann, S., Faissner, A., Holst, A. von An induction gene trap screen in neural stem cells reveals an instructive function of the niche and identifies the splicing regulator Sam68 as a Tenascin-C-regulated target gene. Stem Cells. 26 (9), 2321-2331 (2008).
- Montalbán-Loro, R., et al. TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn. Nat Comm. 10 (1), 1726(2019).
- Gritti, A., et al. Epidermal and fibroblast growth factors behave as mitogenic regulators for a single multipotent stem cell-like population from the subventricular region of the adult mouse forebrain. J Neurosci. 19 (9), 3287-3297 (1999).
- Nieto-Estévez, V., Pignatelli, J., Araúzo-Bravo, M. J., Hurtado-Chong, A., Vicario-Abejón, C. A global transcriptome analysis reveals molecular hallmarks of neural stem cell death, survival, and differentiation in response to partial FGF-2 and EGF deprivation. PLoS One. 8 (1), 53594(2013).
- Vergaño-Vera, E., Méndez-Gómez, H. R., Hurtado-Chong, A., Cigudosa, J. C., Vicario-Abejón, C. Fibroblast growth factor-2 increases the expression of neurogenic genes and promotes the migration and differentiation of neurons derived from transplanted neural stem/progenitor cells. Neuroscience. 162 (1), 39-54 (2009).
- Gabay, L., Lowell, S., Rubin, L. L., Anderson, D. J. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 40 (3), 485-499 (2003).
- Hack, M. A., Sugimori, M., Lundberg, C., Nakafuku, M., Götz, M. Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol Cell Neurosci. 25 (4), 664-678 (2004).
- Bithell, A., Finch, S. E., Hornby, M. F., Williams, B. P. Fibroblast growth factor 2 maintains the neurogenic capacity of embryonic neural progenitor cells in vitro but changes their neuronal subtype specification. Stem Cells. 26 (6), 1565-1574 (2008).
- Pastrana, E., Cheng, L. C., Doetsch, F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci. 106 (15), 6387-6392 (2009).
- Kobayashi, N., et al. Gene delivery to embryonic stem cells. Birth Def Res C. 75 (1), 10-18 (2005).
- Gresch, O., Altrogge, L. Transfection of difficult-to-transfect primary mammalian cells. Meth Mol Biol. 801, 65-74 (2011).
- Maasho, K., Marusina, A., Reynolds, N. M., Coligan, J. E., Borrego, F. Efficient gene transfer into the human natural killer cell line, NKL, using the Amaxa nucleofection system. J Immunol Meth. 284 (1-2), 133-140 (2004).
- von Holst, A., Egbers, U., Prochiantz, A., Faissner, A. Neural stem/progenitor cells express 20 tenascin C isoforms that are differentially regulated by Pax6. J Biol Chem. 282 (12), 9172-9181 (2007).
- Woodard, L. E., Wilson, M. H. piggyBac-ing models and new therapeutic strategies. Trend Biotechnol. 33 (9), 525-533 (2015).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved