A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Generation of Human Chimeric Antigen Receptor Regulatory T Cells
In This Article
Summary
This protocol provides a streamlined workflow to generate and test human chimeric antigen receptor regulatory T cells (CAR Tregs).
Abstract
Chimeric antigen receptor (CAR) T-cell therapy has reshaped the face of cancer treatment, leading to record remission rates in previously incurable hematological cancers. These successes have spurred interest in adapting the CAR platform to a small yet pivotal subset of CD4+ T cells primarily responsible for regulating and inhibiting the immune response, regulatory T cells (Tregs). The ability to redirect Tregs' immunosuppressive activity to any extracellular target has enormous implications for creating cell therapies for autoimmune disease, organ transplant rejection, and graft-versus-host disease. Here, we describe in detail methodologies for bona fide Treg isolation from human peripheral blood, genetic modification of human Tregs utilizing either lentivirus or CRISPR/Cas9-aided knock-in using adeno-associated virus-mediated homologous directed repair (HDR) template delivery, and ex vivo expansion of stable human CAR Tregs. Lastly, we describe the assessment of human CAR Treg phenotypic stability and in vitro suppressive function, which provides insights into how the human CAR Tregs will behave in preclinical and clinical applications.
Introduction
Chimeric antigen receptor (CAR) T-cell therapies have revolutionized the treatment of hematological malignancies, achieving remarkably high remission rates in previously untreatable cancers1,2. Encouraging early results using CAR T cells to treat glioblastoma3,4,5 highlight CAR technology's versatility and future potential to target a wide range of malignancies. As the field explores further applications of CARs, regulatory T cells (Tregs) have emerged as a promising cell type. Tregs play a crucial role in maintaining immune homeostasis, and regulating immune responses through several mechanisms, including sequestering IL-2, secreting immunosuppressive cytokines, and modulating antigen-presenting cells6,7.
With CAR technology, Tregs could be leveraged for the treatment of organ transplant rejection, autoimmune disease, and inflammatory disorders like allergies and asthma6,8,9. CAR Tregs could lead to significant improvements in patient outcomes and quality of life by reducing the use of immunosuppressive drugs, which inhibit the immune system as a whole and are associated with noxious side effects10,11. Preclinical models have shown promising results in translating CAR technology to Tregs, with successful applications in diseases such as type 1 diabetes, multiple sclerosis, graft-versus-host disease, and inflammatory bowel disease9,12,13,14,15. In the clinic, CAR Tregs are currently being explored to prevent solid organ transplant rejection16.
This article presents a detailed methodology for generating human chimeric antigen receptor regulatory T cells (CAR Tregs). This protocol involves isolating Tregs from human peripheral blood and genetically modifying them using techniques such as lentiviral transduction and precise gene knock-in using CRISPR/Cas9 gene editing and adeno-associated virus (AAV) vectors. We also describe the assessment of these engineered Tregs' phenotypic stability and suppressive function, which are crucial steps to validate their therapeutic potential17,18,19. This approach streamlines the design and early testing of CAR Treg therapies, which hold the potential to extend the transformative impact of CAR T-cell therapy to regulate the immune system. By sharing our methodology, we hope to inspire further research and innovation in the burgeoning CAR Treg therapy space9,20.
Protocol
1. Human Treg isolation
- Leukopak processing
- Transfer the contents of the leukopak into a 50 mL conical tube. Add an equal volume of Dulbecco's phosphate-buffered saline (DPBS) +2% fetal bovine serum (FBS) by mixing gently with a pipette.
- Spin at 300 × g for 10 min at room temperature (RT). Carefully aspirate the supernatant. Reconstitute the cell pellet in 2 mL of DPBS + 2% FBS. Add 8 mL of ammonium chloride solution by pipette to the cell suspension at a 4:1 ratio, mix by gentle inversion, and allow the lysis of the remaining red blood cells on ice for 15 min.
- Spin at 300 × g for 10 min at RT. Carefully aspirate the supernatant. Add 30 mL of DPBS + 2% FBS to wash the cells. With the brake off, spin the washed cells at 150 × g for 10 min at RT. Carefully aspirate the supernatant. Resuspend the cell pellet in 30 mL of DPBS + 2% FBS.
- Count the now isolated peripheral blood mononuclear cells (PBMCs) with trypan blue at a 1:1 ratio.
- Due to the high cell concentration, initially dilute 10 µL of cells 1:100 with DPBS; then, mix 10 µL of the 1:100 diluted cells with 10 µL of trypan blue solution for that final 1:1 dilution. When using an automated cell counter, correct the cell count to reflect the 200-fold dilution by multiplying the reported concentration by 100, as most counters assume a 2-fold dilution. Anticipate yielding 1-2.5 × 109 PBMC from a 1/10 leukopak.
- CD4+ T cell isolation (negative selection)
- Spin down 108-109 PBMCs at 500 × g for 5 min at RT and resuspend in Cell Separation Buffer (DPBS + 1 mM EDTA + 2% FBS) at 5 × 107 cells/mL.
NOTE: To obtain enough human Tregs for genetic modification, we recommend starting with 1 × 109 PBMCs. - Perform magnetic separation according to the manufacturer's instructions for the CD4+ T cell enrichment kit.
- Determine the amount of CD4+ T cells isolated by counting with trypan blue (10 µL of cells + 10 µL of trypan blue).
- Spin down 108-109 PBMCs at 500 × g for 5 min at RT and resuspend in Cell Separation Buffer (DPBS + 1 mM EDTA + 2% FBS) at 5 × 107 cells/mL.
- CD8+ T cell isolation (negative selection)
- Spin 5 × 107 PBMCs at 500 × g for 5 min at RT. Resuspend in Cell Separation Buffer (DPBS + 1 mM EDTA + 2% FBS) at 5 × 107 cells/mL.
NOTE: We recommend starting with 5 × 107 PBMC to obtain 2-5 × 106 CD8+ T cells. - Perform magnetic separation according to the manufacturer's instructions for the CD8+ T cell enrichment kit.
- Determine the number of CD8+ T cells isolated by counting with trypan blue (10 µL of cells + 10 µL of trypan blue).
- Spin 5 × 107 PBMCs at 500 × g for 5 min at RT. Resuspend in Cell Separation Buffer (DPBS + 1 mM EDTA + 2% FBS) at 5 × 107 cells/mL.
- Treg fluorescence-assisted cell sorting (FACS)
- Isolate CD4+ T cells as described in step 1.2 and store them overnight in DPBS with 2% FBS at 4 °C (minimal loss in cell number and viability) for FACS the following day. Determine the number of CD4+ T cells from step 1.2 by counting with trypan blue (10 µL of cells + 10 µL of trypan blue).
- Spin CD4+ cells at 500 × g for 5 min. Reconstitute cells in 200 µL of DPBS.
- Per 1 × 106 cells, add 1 µL of anti-human CD4 FITC, 1 µL of anti-human CD25 APC, and 1 µL of anti-human CD127 PE. Gently vortex and place in the 4 °C dark fridge for 30 min.
- Wash the cells with 10 mL of DPBS with 2% FBS. Spin at 500 × g for 5 min. Gently resuspend the stained cells at 1.5 × 107 cells/mL in DPBS with 2% FBS. This is the recommended cell concentration to sort via fluorescence-activated cell sorting (FACS).
- Pass the stained cell suspension through a 40 µm filter cap into FACS tubes, and then keep the tubes on ice.
- Prepare 15 mL collection tubes containing 3 mL of RPMI10 medium and place on ice.
NOTE: This medium consists of RPMI1640 basal medium, 10% FBS, 1x penicillin-streptomycin, 1x L-alanyl-L-glutamine, 1x non-essential amino acids, 1x sodium pyruvate, and 1x HEPES. - Sort CD4+CD25highCD127- regulatory T cells (Tregs) and CD4+CD25lowCD127+ conventional T cells (Tconv) using FACS as shown in Figure 1A.
- Determine cell yield and viability; then proceed with downstream analysis or T cell activation.
2. T cell activation
- Count isolated T cells with trypan blue (10 µL of cells + 10 µL of trypan blue).
- Wash 25 µL of anti-CD3/CD28 beads (106 beads) for every 1 × 106 T cells obtained, maintaining a 1:1 ratio of beads to T cells. Wash anti-CD3/CD28 beads by adding DPBS, incubating on a magnet for 3 min, and removing the now diluted anti-CD3/CD28 bead buffer, which can be toxic to cells.
- Remove the tube from the magnet and resuspend the washed beads in RPMI10 medium to have 1 × 106 beads/1 mL of RPMI10.
- Resuspend T cells with anti-CD3/CD28 beads in RPMI10 medium at a concentration of 1 × 106 T cells/mL. Then, add 1,000 IU/mL of IL-2 to Treg cells, 100 IU/mL of IL-2 to CD4+ Tconv cells, and 300 IU/mL of IL-2 to CD8+ T cells.
- Culture T cells at 1 × 106 cells per well of a 24-well plate with different amounts of anti-CD3/CD28 beads and IL-2. Place the 24-well plate in a 37 °C, 5% CO2 tissue culture incubator.
- Expand activated T cells for 9-12 days in the presence of IL-2 until being used for experiments or re-activation with anti-CD3/CD28 beads for additional expansion. Replace media every 2-3 days and add IL-2 every time fresh medium is added. Keep the cell density of the Tregs at 5 × 105 to 1 × 106 cells per mL during expansion by counting the cells or by visual inspection before splitting.
3. Human Treg lentiviral transduction
- Resuspend and count Tregs 48 h after activation. Spin at 500 ×g for 5 min at RT. Resuspend Tregs in RPMI10 at 1.25 × 106 cells/mL with 1,000 IU/mL of IL-2.
- Prewarm the centrifuge to 32 °C.
- Thaw the lentivirus-containing CAR construct on ice.
NOTE: We store single-use lentivirus aliquots with enough lentivirus to transduce 2.5 × 105 cells at an MOI of 1 at -80 °C. - Add each lentivirus aliquot to 2.5 × 105 Tregs in 200 µL in a microcentrifuge tube, each tube indicating a reaction. Spinoculate at 1,000 × g for 1 h at 32 °C.
- Move each 200 µL reaction to a 24-well plate. Make sure the media covers the entire well. Incubate the 24-well plate with the transduced Tregs in a tissue culture incubator overnight (16-18 h). Top up each well to 2 mL with RPMI10 medium with the final IL-2 concentration being 1,000 IU/mL.
- Continue expanding transduced Tregs 9-12 days post activation by splitting the cells and supplementing with fresh prewarmed RPMI10 and IL-2 as needed (every 2-3 days). The ideal concentration of cells is 5 × 105-1 × 106 cells/mL.
- Assess gene modification efficiency using flow cytometry as shown in Figure 2.
NOTE: Our lentiviral CAR constructs contain a Myc-tag on the N-terminus of the CAR gene and a GFP reporter gene linked to the CAR gene by a 2A peptide, allowing for quantification of transduction efficiency without antibody staining (GFP) and confirmation of CAR surface expression (Myc-tag). We assess gene modification efficiency 5 days after transduction. Transduction efficiencies can vary between 30% and 70% depending on the donor and the CAR construct utilized. CAR+ cells can be sorted if desired. - Ensure that the modified Tregs complete their activation cycle (9-12 days) and rest 24 h in the absence of IL-2 before being used in any experiments.
4. CRISPR/Cas9-mediated gene knock-in in human Tregs
- Resuspend Tregs and transfer to a 15 mL conical tube 48 h after activation. Incubate the cell suspension in a magnet for 3 min.
- While in the magnet, transfer the cells in the medium via pipette to a new tube. The anti-CD3/CD28 beads will remain attached to the tube wall. Allow the debeaded Tregs to rest in RPMI10 for 2 h after bead removal to recover from the immediate stress of debeading, enhancing future transduction efficiency and recovery from electroporation.
- Count the debeaded Tregs with trypan blue.
- Prewarm reduced-serum medium without FBS to 37 °C.
- Prepare a 6-well plate with 2.5 mL of RPMI10 medium without antibiotics (penicillin-streptomycin) and with 1,000 IU/mL IL-2 per well. Prewarm the plate to 37 °C in a tissue culture incubator.
- Thaw the adeno-associated virus (AAV)-containing CAR homology-directed repair (HDR) template on ice.
NOTE: We store single-use aliquots with enough AAV to infect 4 × 106 cells at an MOI of 20,000 at -80 °C. - Spin Tregs at 500 × g for 5 min. After decanting the supernatant, resuspend cells in prewarmed reduced-serum medium at 4 × 107 cells/mL.
- Aliquot cells in 100 µL in low protein binding 1.5 mL centrifuge tubes. Add CAR AAV at an MOI of 20,000 to each sample and resuspend. Incubate the reaction tubes in the tissue culture incubator for 1 h.
- During the 1 h incubation, prepare and assemble CRISPR/Cas9 ribonucleoprotein (RNP) complexes by slowly adding 8.3 µL of Cas9 protein (1 mg/mL stock) to 2.5 µL of sgRNA targeting the TRAC gene locus (100 µM stock) for a molar ratio of Cas9 to sgRNA of 1:1 and a total RNP volume of 10.8 µL per sample. Gently pipet up and down to mix. Incubate the RNP mixture for 15 min at 37°C in the tissue culture incubator.
NOTE: RNP complexes can remain at RT afterward until ready to use. - Fill a fresh electroporation tube (E tube) with 3 mL of high-osmolarity electroporation buffer. Insert the filled E Tube into the pipette station of the electroporation system until a click is heard. Set electroporation conditions to 2,200 V, 20 ms, 1 pulse in the electroporation system.
- When the 1 h incubation with AAV is complete, spin the cells with AAV at 300 × g for 5 min at RT. Carefully aspirate the supernatant and resuspend the cell pellet in 100 µL of the cell resuspension buffer provided by the electroporation system per sample.
NOTE: Work quickly and avoid leaving cells in the buffer for longer than 15 min. Avoid creating bubbles. - Add 10.8 µL of RNP complex per sample. Mix well with a pipette without creating bubbles.
- Insert a 100 µL electroporation tip by pushing the pipette to its second stop to open the clamp. Position the pipette's top head into the electroporation tip until the clamp securely engages with the piston's mounting stem. Gradually release the button while maintaining downward pressure on the pipette to ensure the tip fits snugly without any gaps.
- Press the pipette to the first stop and immerse the electroporation tip into the cell-RNP mixture. Gently pull up the sample into the pipette without any bubbles.
NOTE: No bubbles can be present inside the tip during electroporation. - Insert the pipette with the mounted electroporation tip containing the sample vertically into the E Tube until a click sound is heard. Avoid creating bubbles.
- Confirm that optimal settings for human Tregs have been entered (2,200 V, 20 ms, 1 Pulse) and press start on the touchscreen to electroporate the cells.
- Wait for the touchscreen to display Complete upon completion of the electroporation. Gently remove the pipette and immediately transfer the sample into the prepared 6-well plate containing 2.5 mL of prewarmed, antibiotic-free RPMI10 medium with 1,000 IU/mL of IL-2 per well. Repeat with remaining samples. Gently rock the plate in linear motions (left to right, top to bottom) to ensure even distribution of cells in each well and place it in the tissue culture incubator.
NOTE: Reusing an electroporation tip is acceptable up to 3x if the same cells and RNP complex are in use. Allow a 30 min recovery period before counting the cells or disturbing them in any way such as adding an HDR enhancer if desired. - The following day, 16-18 h later, replace the media with antibiotic-containing media, count the electroporated Tregs with trypan blue, and culture at 106 cells/mL with 1,000 IU/mL IL-2. Continue expanding electroporated Tregs by splitting the cells and supplementing fresh RPMI10 and IL-2 as described in step 2.6.
- Keep samples on ice for analysis by flow cytometry.
- Set up single-color compensation controls and apply to the experiment file.
- Read the unstained sample first to adjust FSC and SSC gains so that the lymphocyte population is in the middle of the 'All events' dot plot.
- Set up gating strategy of lymphocytes into non-debris/single cells into viable cells into CD4+ cells, as displayed in Figure 3C.
- Assess gene modification efficiency using flow cytometry.
NOTE: Our AAV CAR constructs contain, in addition to TRAC locus homology arms, a truncated epidermal growth factor receptor (EGFRt) reporter gene linked to the CAR gene by a 2A peptide as shown in Figure 3. Gene knock-in efficiency is determined by surface staining for CD3, the loss of which indicates loss of TCR surface expression and, hence, successful targeting of the TRAC locus with CRISPR/Cas9, and EGFRt, whose expression indicates successful integration of the CAR transgene. CAR knock-in Tregs are CD3-EGFRt+ cells. We assess gene modification efficiency 5 days after electroporation. - Ensure that modified Tregs complete their activation cycle (9-12 days) and rest 24 h in the absence of IL-2 before being used in any experiments.
5. Human CAR Treg activation
- Co-culture setup (Day 0)
- Collect the CAR antigen-expressing target cell line into a conical tube.
NOTE: We use K562 cells, a human myelogenous leukemia cell line that lacks HLA, CD80, and CD86 expression as target cells for CAR Treg activation. Parental K562 cells are used as a negative control and CAR antigen-expressing K562 are used to activate CAR Tregs17,21. - Irradiate target cell lines with 4,000 rad in a Cesium-137 or X-ray irradiator. In case of lack of access to an irradiator, perform mitomycin C treatment to stop cell proliferation while maintaining surface antigen expression in target cells22.
- If not done so already, debead the Tregs by resuspending the Tregs bound to the anti-CD3/CD28 beads and transferring them to a 15 mL conical tube. Incubate the cell suspension in a magnet for 3-5 min. While still in the magnet, transfer the cells in the medium via pipette to a new tube; the anti-CD3/CD28 beads will remain attached to the tube wall.
- Determine the irradiated target cell and debeaded CAR Treg concentrations with trypan blue.
- Spin irradiated target cells and CAR Tregs at 500 × g for 5 min. Resuspend with prewarmed RPMI10 at 106 cells/mL in their separate tubes. Add IL-2 to CAR Tregs for a concentration of 2,000 IU/mL.
NOTE: This amount of IL-2 will be diluted 2-fold once the CAR Tregs are combined with the target cells for a final IL-2 concentration of 1,000 IU/mL. - Co-culture 1 × 105 CAR Tregs (100 µL) with 1 × 105 (100 µL) CAR antigen-negative, irradiated target cells (negative control for activation), 1 × 105 (100 µL) CAR antigen-positive, irradiated target cells (experiment), or 2.5 µL of anti-CD3/CD28 beads (positive control for activation) and 97.5 µL of RPMI10 medium in a 96-well round bottom plate. Place the plate in a 37 °C, 5% CO2 tissue culture incubator for 48 h.
NOTE: The final volume per well is 200 µL. Ensure that each condition has three replicates.
- Collect the CAR antigen-expressing target cell line into a conical tube.
- Flow cytometry readout
- Unless using a plate reader, resuspend the contents of each well from the 96 well round-bottom plate and transfer them into a FACS tube. Spin at 500 x g for 5 min. Decant the supernatant and gently vortex the cell pellet.
- Prepare flow cytometry antibody master mix: 100 µL/sample containing DPBS, anti-human CD4 PE/Cy7 1:200, anti-human CD71 PE 1:100, and Ghost Viability Dye BV510 1:2000.
NOTE: This panel works well if the CAR reporter gene is GFP. If a reporter gene encodes a surface protein that requires antibody staining, such as EGFRt, a FITC-conjugated antibody can be used, for example. - Pipette 100 µL of the antibody master mix into each sample, gently mix, and incubate for 30 min in the 4 °C fridge.
- Wash with 500 µL of DPBS by spinning at 500 × g for 5 min. Resuspend the cell pellet in 200 µL of DPBS.
- Keep samples on ice for analysis by flow cytometry.
- Set up single-color compensation controls and apply to experiment flow file.
- Read unstained samples to adjust SSC and FSC gains so that lymphocyte population is in the middle of the 'All events' dot plot.
- Set up gating strategy of lymphocytes into non-debris/single cells into viable cells into CD4+ cells, as displayed in Figure 3C (Different fluorophores used).
- Read samples at an event rate of roughly 1,500 events/s to assess Treg activation.
NOTE: The expected outcome is upregulation of CD71 surface expression in CAR Tregs in the presence of CAR antigen as shown in Figure 4. If upregulation is not seen in the presence of CAR antigen, then tonic signaling could be present.
6. Human CAR Treg stability
- Co-culture setup and expansion (Days 0-9)
- Set up co-cultures to activate CAR Tregs as described in step 5.1.
- After 48 h, transfer co-culture from each 96-well round-bottom plate well into a 24-well plate well containing prewarmed 2 mL of RPMI10 with 1,000 IU/mL IL-2 to allow for cell expansion.
- Add fresh prewarmed RPMI10 with 1,000 IU/mL IL-2 and split into additional 24-well plates as needed.
- Flow cytometry readout
- Resuspend and transfer the contents of each replicate into a 15 mL or 50 mL conical tube. Determine cell concentrations.
- Transfer between 5 × 105 and 1 × 106 cells to a FACS tube for each replicate. Spin at 500 × g for 5 min.
- Prepare flow cytometry antibody master mix for surface staining: 100 µL/sample containing DPBS, anti-human CD4 PE/Cy7 1:200, anti-human CD25 APC 1:200, and Ghost Viability Dye BV510 1:2000. Include FITC-conjugated antibody for CAR reporter protein if needed.
- Decant the supernatant from step 6.2.2 and gently vortex the cell pellet. Pipette 100 µL of the surface-staining antibody master mix to each FACS tube. Briefly vortex and place in a 4 °C fridge for 30 min in the dark.
- Using a transcription factor staining buffer set, prepare Fixation/Permeabilization buffer by adding 3 volumes of Fixation/Permeabilization diluent to 1 volume of Fixation/Permeabilization concentrate. Each sample requires 100 µL of Fixation/Permeabilization buffer.
- Wash surface-stained cells with 500 µL of DPBS. Spin at 500 × g for 5 min and decant the supernatant. Pipette 100 µL of prepared Fixation/Permeabilization buffer to each tube. Briefly vortex and allow fixation to occur at 4 °C for 30-60 min in the dark.
- Using a transcription factor staining buffer set, prepare 1x Permeabilization buffer by adding 9 volumes of distilled water to 1 volume of Permeabilization Buffer 10x concentrate. Each sample requires 1,000 µL of Permeabilization buffer for washing and 100 µL of Permeabilization buffer for staining with antibodies targeting intracellular proteins.
- Wash the fixed/permeabilized cells by adding 500 µL of 1x Permeabilization buffer. Spin at 500 × g for 5 min at RT.
- Prepare intracellular staining antibody master mix with 1x Permeabilization buffer, anti-human FOXP3 eFluor 450 1:50, anti-human HELIOS PE 1:50, and anti-human CTLA-4 PerCP-e710 1:50. Each sample will require 100 µL of antibody master mix.
NOTE: This panel works if the CAR reporter gene is GFP or if the CAR reporter protein (e.g., EGFRt) is stained with a FITC-conjugated antibody. - Decant the supernatant from step 6.2.8. Add 100 µL of the intracellular staining antibody master mix, briefly vortex, and incubate at RT for 30 min in the dark.
- Wash the stained fixed/permeabilized cells by adding 500 µL of 1x Permeabilization buffer. Spin at 500 × g for 5 min. Decant the supernatant, resuspend the cell pellet in 300 µL of DPBS, and store it on ice.
- Analyze by flow cytometry as shown in Figure 5. The anticipated result is that most CAR Tregs will be FOXP3+HELIOS+ cells. Use CD4+ Tconv cells as a negative control for FOXP3 and HELIOS staining.
7. Human CAR Treg suppression
- Responder T cell (Tresp) cell trace dye staining and overnight activation
- Collect the expanded CAR Treg cells, freshly isolated CD4+ Tconv cells, and freshly isolated CD8+ T cells into separate 15 mL conical tubes and debead anti-CD3/CD28 beads if not done so already.
- Prepare irradiated CAR antigen-expressing target cells as described in step 5.1.
- Determine non-activated T cell and irradiated target cell concentrations.
- Combine 5 × 106 CD4+ Tconv cells with 5 × 106 CD8+ T cells (1:1 ratio). These are the responder T cells (Tresp) to be inhibited by CAR Tregs in the assay. Spin Tresp cells at 500 × g for 5 min at RT. Carefully aspirate the supernatant and resuspend Tresp cells in 1 mL of DPBS.
- Add 1 mL of 5 mM CellTrace Violet (CTV) dye reconstituted in DMSO to the 107 Tresp cells in 1 mL of DPBS for a final concentration of 5 µM CTV. Place in a water bath at 37 °C for 20 min. At 10 min, gently vortex to redistribute the settled CTV.
- Wash with 9 mL of prewarmed RPMI10 complete medium. Spin at 500 × g for 5 min. Resuspend in 5 mL of prewarmed RPMI10 complete medium.
- Determine CTV-labeled Tresp cell concentration. Add 5 × 104 CTV-labeled non-activated Tresp cells in 200 µL of RPMI10 medium in 3-6 wells of a 96-well round-bottom plate as minimum proliferation controls.
- Activate CTV-labeled Tresp cells with anti-CD3/CD28 beads at a 1:10 bead to Tresp cell ratio without IL-2 in RPM10 medium. Dispense 106 CTV-labeled Tresp cells in 1 mL of RPMI10 medium per well of a 24-well plate in the tissue culture incubator overnight.
- In parallel, obtain 106 CAR+ Tregs. This amount covers triplicates of four CAR Treg:Tresp ratios, 1:1, 1:2, 1:4, 1:8, at 5 × 104 target cells per 96-well round-bottom plate well, as 5 × 104 × 3 + 2.5 × 104 × 3 + 1.25 × 104 × 3 + 0.625 × 104 × 3 = 2.81 × 105 CAR Tregs, across two CAR Treg activation conditions: irradiated CAR antigen-negative target cells (negative control, no activation), and irradiated CAR antigen-positive target cells (experiment, CAR activation).
- Combine 3 × 105 CAR Tregs with 3 × 105 irradiated CAR antigen-negative cells (1:1 ratio) and 3 × 105 CAR Tregs with 3 × 105 irradiated CAR antigen-positive target cells (1:1 ratio), in two separate 15 mL conical tubes and spin at 500 × g for 5 min at RT. Carefully aspirate the supernatant.
- Resuspend the pellet in each tube from step 7.1.10 in 600 µL of prewarmed RPMI10 medium. This results in 200 µL for each of the 1:1 Treg: Tresp triplicates. Perform serial dilution on a 96-well round-bottom plate as follows:
- From the prepared 600 µL of Treg + target cell suspension, pipette 200 µL of cell suspension into each of the 3 1:1 ratio wells.
- Add 100 µL of prewarmed RPMI10 complete medium into each of the empty 1:2, 1:4, and 1:8 ratio wells.
- For each of the triplicates, pipette 100 µL of the cell suspension from the 1:1 wells into the associated 1:2 ratio wells.
- For each of the triplicates, pipette 100 µL of the cell suspension, from the 1:2 wells, into the associated 1:4 ratio wells.
- For each of the triplicates, pipette 100 µL of the cell suspension, from the 1:4 wells, into the associated 1:8 ratio wells.
- Pipette the remaining 100 µL of the cell suspension from the 1:8 ratio wells into a waste container.
NOTE: Each well should contain 100 µL of the cell suspension with either 5 × 104 (1:1), 2.5 × 104 (1:2), 1.25 × 104 (1:4), or 0.625 × 104 (1:8) CAR Tregs and an equal number of irradiated target cells.
- CAR Treg and Tresp cell co-incubation
- After 16-18 h post activation, collect activated Tresp cells from the 24-well plate into a conical tube and remove anti-CD3/CD28 beads via magnet.
- Determine debeaded CTV-labeled Tresp cell counts.
- Wash and spin 2 × 106 Tresp cells at 500 × g for 5 min. Carefully aspirate the supernatant. Resuspend the cells in 4 mL of RPMI10 complete medium.
- Add 100 µL of the T cell suspension (5 × 104 T cells) to each well with CAR Tregs, as well as to 3-6 wells with 100 µL of RPMI10 medium alone for maximum proliferation controls. Place the plate in a 37 °C, 5% CO2 tissue culture incubator for 72 h.
NOTE: The 96-well round-bottom plate now has 3-6 wells with non-activated Tresp cells alone (minimum proliferation control), 3-6 wells with activated Tresp cells alone (maximum proliferation control) and activated Tresp cells in the presence of decreasing numbers of CAR Tregs.
- Flow cytometry readout
- Resuspend and transfer the contents of each 96-well round bottom plate well into a labeled FACS tube. Alternatively, transfer to a 96-well V-bottom plate if a flow cytometer with plate reading mode is available. Spin at 500 × g for 5 min.
- In the meantime, prepare antibody master mix with DPBS, anti-human CD4 PE/Cy7 1:200, and anti-human CD8 PerCP 1:200. Each sample will require 100 µL of antibody master mix.
- Gently vortex the cell pellet. Pipette 100 µL of antibody master mix into each FACS tube. Briefly vortex and incubate at 4 °C for 30 min in the dark.
- Wash with 500 µL of DPBS. Wash 2x with 100 µL of DPBS if using a 96-well V-bottom. Spin at 500 × g for 5 min. Decant the supernatant, resuspend the cell pellet in 200 µL of DPBS, and store tubes on ice in the dark.
- Analyze by flow cytometry as shown in Figure 6. The expected outcome is non-activated Tresp cells alone (minimum proliferation) to display a uniform high CTV fluorescence peak, activated Tresp cells alone displaying multiple peaks of CTV intensity, one corresponding to each cell division (maximum proliferation), and activated CD4+ and CD8+ T cells in the presence of activated CAR Tregs displaying a reduction in the number and height of CTV peaks, hence, in proliferation.
- Calculate Treg cell-mediated suppression as follows:
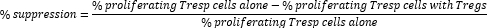
NOTE: If using FlowJo software, it is also possible to use cell proliferation modeling to calculate the division index (DI) for each sample and calculate percentage suppression using DI instead of percentage of proliferating cells in the formula above.
Results
The protocol described here provides a streamlined and standardized pipeline to assess new chimeric antigen receptor (CAR) constructs in human regulatory T cells (Tregs), with the aim of creating living therapeutics for autoimmune disease, graft-versus-host disease, organ transplant rejection, and allergy. Figure 1 depicts how we obtain highly pure human Tregs from peripheral blood using FACS (Figure 1A), as evaluated by their high levels of the Treg lineage tra...
Discussion
This protocol provides a streamlined and comprehensive methodology for generating and evaluating human chimeric antigen receptor regulatory T cells (CAR Tregs). The success of CAR technology in treating hematological cancers has inspired its application to the immunosuppressive subset of T cells, Tregs. Unlike conventional T cells, Tregs inhibit immune responses, offering potential treatments for conditions resulting from unwanted immunity, such as autoimmune disease, organ transplant rejection, graft-versus-host disease...
Disclosures
LMRF is an inventor and has received royalties from patents on engineered immune cells and consults for Guidepoint Global and McKesson. The remaining authors declare no competing interests.
Acknowledgements
LMRF is funded by Human Islet Research Network (HIRN) Emerging Leader in Type 1 Diabetes grant U24DK104162-07, American Cancer Society (ACS) Institutional Research Grant IRG-19-137-20, South Carolina Clinical and Translational Research (SCTR) Pilot Project Discovery Grant 1TL1TR001451-01, Diabetes Research Connection (DRC) Grant IPF 22-1224, and Swim Across America Grant 23-1579. RWC is supported by the Cellular, Biochemical and Molecular Sciences training grant T32GM132055 and Hollings Cancer Center Lowvelo Graduate Fellowship. This study was supported in part by the Flow Cytometry and Cell Sorting Shared Resource, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313). Special thanks to Dr. Qizhi Tang at the University of California, San Francisco (UCSF) for kindly gifting the CAR mutant plasmids.
Materials
| Name | Company | Catalog Number | Comments |
| Adeno-associated virus (AAV) | Charles River Laboratories | ||
| CAR target-expressing K562 cells | e.g., CD19-K562 | ||
| Cesium-137 irradiator | |||
| Anti-human CD8 PerCP (clone SK1) | Biolegend | 344708 | |
| Anti-human CD4 PE/Cy7 (clone SK3) | Biolegend | 344612 | |
| DynaMag-15 magnet | ThermoFisher | 12301D | |
| Ghost BV510 viability dye | TONBO | 13-0870-T100 | |
| K562 cells | American Type Culture Collection | CCL-243 | |
| 0.5 M EDTA, pH 8.0 | Gibco | 15575020 | |
| 1 M HEPES | Gibco | 15630080 | |
| Ammonium chloride solution | STEMCELL Technologies | 7850 | |
| Anti-human CD127 PE (clone hIL-7R-M21) | BD Biosciences | 557938 | |
| Anti-human CD25 APC (clone BC96) | Biolegend | 302610 | |
| Anti-human CD4 FITC (clone SK3) | Biolegend | 344604 | |
| Anti-human CD71 PE (clone SK1) | Biolegend | 334106 | |
| Anti-human CD8 PerCP (clone SK1) | Biolegend | 344707 | |
| Anti-human CTLA-4 PerCP-e710 | ThermoFisher | 46-1529-42 | |
| Anti-human EGFR APC (clone AY13) | Biolegend | 352905 | |
| Anti-human FOXP3 eFluor 450 | ThermoFisher | 48-4776-42 | |
| Anti-human HELIOS PE | Biolegend | 137216 | |
| Ca2+ and Mg2+ free Dulbecco’s Phosphate Buffered Saline (DPBS) | Gibco | 14190144 | |
| Cell counter (TC20 Automated Cell Counter) | Bio-Rad | 1450102 | |
| Cell Counting Slides | Bio-Rad | 1450016 | |
| CellTrace Violet Cell Proliferation Kit | ThermoFisher | C34571 | |
| DNA LoBind Tubes | Eppendorf | 22431021 | |
| Easy 50 EasySep magnet | STEMCELL Technologies | 18002 | |
| EasySep Human CD4+ T cell Enrichment Kit | STEMCELL Technologies | 19052 | |
| EasySep Human CD8+ T cell Enrichment Kit | STEMCELL Technologies | 19053 | |
| EasySep magnet | STEMCELL Technologies | 18000 | |
| eBioscience Foxp3 transcription factor staining buffer set | ThermoFisher | 00-5523-00 | |
| Falcon Round-Bottom Polystyrene Test Tubes with Cell Strainer Snap Cap, 5 mL | Fisher Scientific | 08-771-23 | 40μm |
| Fetal Bovine Serum (FBS) | Gibco | 26140079 | |
| Flow cytometer | Beckman Coulter | CytoFLEX LX U3-V5-B3-Y5-R3-I2 | |
| Fluorescence-activated cell sorter | BD Biosciences | FACS Aria III Cell Sorter | |
| GlutaMAX | Gibco | 35050061 | |
| Human CD3/28 T Cell Expansion and Activation Dynabeads | Gibco | 11131D | |
| Invitrogen Neon Transfection System | ThermoFisher | 10431915 | |
| Invitrogen Neon Transfection System 100 μL Kit | ThermoFisher | 10114334 | |
| Lentivirus | VectorBuilder | ||
| MEM Non-Essential Amino Acids Solution | Gibco | 11140050 | |
| Myc Tag antibody A647 (clone 9B11) | Cell Signaling Technologies | 2233S | |
| Opti-MEM I Reduced Serum Medium | ThermoFisher | 31985062 | |
| Penicilin-Streptomycin solution | Gibco | 15140122 | |
| Recombinant human interleukin-2 (rhIL-2) | Peprotech | 200-02 | |
| RPMI 1640 medium, no glutamine | Gibco | 11875093 | |
| Sodium pyruvate | Gibco | 11360070 | |
| Spectral Flow Cytometer | Cytek | Northern Lights | |
| TRAC gRNA | Synthego | Sequence (CAGGGTTCTGGATATCTGT) | |
| TrueCut Cas9 Protein v2 | ThermoFisher | A36496 | |
| Trypan Blue solution | Sigma | T8154-100ML | |
| 1/10 Leukopak | STEMCELL Technologies | 200-0092 | 1-2 billion PBMC |
References
- Zhang, X., Zhu, L., Zhang, H., Chen, S., Xiao, Y. CAR-T cell therapy in hematological malignancies: current opportunities and challenges. Front Immunol. 13, 927153 (2022).
- Cappell, K. M., Kochenderfer, J. N. Long-term outcomes following CAR T cell therapy: what we know so far. Nat Rev Clin Oncol. 20 (6), 359-371 (2023).
- Choi, B. D., et al. Intraventricular CARv3-TEAM-E T cells in recurrent glioblastoma. N Engl J Med. 390 (14), 1290-1298 (2024).
- Brown, C. E., et al. Locoregional delivery of IL-13Ralpha2-targeting CAR-T cells in recurrent high-grade glioma: a phase 1 trial. Nat Med. 30 (4), 1001-1012 (2024).
- Bagley, S. J., et al. Intrathecal bivalent CAR T cells targeting EGFR and IL13Ralpha2 in recurrent glioblastoma: phase 1 trial interim results. Nat Med. 30 (5), 1320-1329 (2024).
- Ghobadinezhad, F., et al. The emerging role of regulatory cell-based therapy in autoimmune disease. Front Immunol. 13, 1075813 (2022).
- Sakaguchi, S., Yamaguchi, T., Nomura, T., Ono, M. Regulatory T cells and immune tolerance. Cell. 133 (5), 775-787 (2008).
- Sakaguchi, S., et al. Regulatory T cells and human disease. Annu Rev Immunol. 38, 541-566 (2020).
- Ferreira, L. M. R., Muller, Y. D., Bluestone, J. A., Tang, Q. Next-generation regulatory T cell therapy. Nat Rev Drug Discov. 18 (10), 749-769 (2019).
- Rosenblum, M. D., Gratz, I. K., Paw, J. S., Abbas, A. K. Treating human autoimmunity: current practice and future prospects. Sci Transl Med. 4 (125), 125sr121 (2012).
- Sawitzki, B., et al. Regulatory cell therapy in kidney transplantation (The ONE Study): a harmonised design and analysis of seven non-randomised, single-arm, phase 1/2A trials. Lancet. 395 (10237), 1627-1639 (2020).
- Spanier, J. A., et al. Tregs with an MHC class II peptide-specific chimeric antigen receptor prevent autoimmune diabetes in mice. J Clin Invest. 133 (18), e168601 (2023).
- Muller, Y. D., et al. Precision engineering of an anti-HLA-A2 chimeric antigen receptor in regulatory T cells for transplant immune tolerance. Front Immunol. 12, 686439 (2021).
- MacDonald, K. G., et al. Alloantigen-specific regulatory T cells generated with a chimeric antigen receptor. J Clin Invest. 126 (4), 1413-1424 (2016).
- Boardman, D. A., et al. Flagellin-specific human CAR Tregs for immune regulation in IBD. J Autoimmun. 134, 102961 (2023).
- Schreeb, K., et al. Study design: human leukocyte antigen Cclass I molecule A(*)02-chimeric antigen receptor regulatory T cells in renal transplantation. Kidney Int Rep. 7 (6), 1258-1267 (2022).
- Zimmerman, C. M., Robino, R. A., Cochrane, R. W., Dominguez, M. D., Ferreira, L. M. R. Redirecting human conventional and regulatory T cells using chimeric antigen receptors. Methods Mol Biol. 2748, 201-241 (2024).
- Tang, Q., et al. Selective decrease of donor-reactive T(regs) after liver transplantation limits T(reg) therapy for promoting allograft tolerance in humans. Sci Transl Med. 14 (669), eabo2628 (2022).
- Bender, C., et al. A phase 2 randomized trial with autologous polyclonal expanded regulatory T cells in children with new-onset type 1 diabetes. Sci Transl Med. 16 (746), eadn2404 (2024).
- Ferreira, L. M. Conference report: Advanced Therapies Week 2023. Regen Med. 18 (4), 297-299 (2023).
- Cochrane, R. W., et al. How to test human CAR T cells in solid tumors, the next frontier of CAR T cell therapy. Methods Mol Biol. 2748, 243-265 (2024).
- Roy, A., Krzykwa, E., Lemieux, R., Neron, S. Increased efficiency of gamma-irradiated versus mitomycin C-treated feeder cells for the expansion of normal human cells in long-term cultures. J Hematother Stem Cell Res. 10 (6), 873-880 (2001).
- Eyquem, J., et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 543 (7643), 113-117 (2017).
- Negrini, M., Wang, G., Heuer, A., Bjorklund, T., Davidsson, M. AAV Production everywhere: a simple, fast, and reliable protocol for in-house AAV vector production based on chloroform extraction. Curr Protoc Neurosci. 93 (1), e103 (2020).
- Velasco Cardenas, R. M., et al. Harnessing CD3 diversity to optimize CAR T cells. Nat Immunol. 24 (12), 2135-2149 (2023).
- Boomer, J. S., Green, J. M. An enigmatic tail of CD28 signaling. Cold Spring Harb Perspect Biol. 2 (8), a002436 (2010).
- Suhoski, M. M., et al. Engineering artificial antigen-presenting cells to express a diverse array of co-stimulatory molecules. Mol Ther. 15 (5), 981-988 (2007).
- Fung, V. C. W., Rosado-Sanchez, I., Levings, M. K. Transduction of human T cell subsets with lentivirus. Methods Mol Biol. 2285, 227-254 (2021).
- Bailey-Bucktrout, S. L., et al. Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity. 39 (5), 949-962 (2013).
- Nakagawa, H., et al. Instability of Helios-deficient Tregs is associated with conversion to a T-effector phenotype and enhanced antitumor immunity. Proc Natl Acad Sci U S A. 113 (22), 6248-6253 (2016).
- Dawson, N. A. J., et al. Functional effects of chimeric antigen receptor co-receptor signaling domains in human regulatory T cells. Sci Transl Med. 12 (557), eaaz3866 (2020).
- Rana, J., et al. CAR- and TRuC-redirected regulatory T cells differ in capacity to control adaptive immunity to FVIII. Mol Ther. 29 (9), 2660-2676 (2021).
- Battaglia, M., Stabilini, A., Tresoldi, E. Expanding human T regulatory cells with the mTOR-inhibitor rapamycin. Methods Mol Biol. 821, 279-293 (2012).
- Brady, B. L., Steinel, N. C., Bassing, C. H. Antigen receptor allelic exclusion: an update and reappraisal. J Immunol. 185 (7), 3801-3808 (2010).
- Samarasinghe, S., et al. Functional characterization of alloreactive T cells identifies CD25 and CD71 as optimal targets for a clinically applicable allodepletion strategy. Blood. 115 (2), 396-407 (2010).
- Voss, K., et al. FOXP3 protects conventional human T cells from premature restimulation-induced cell death. Cell Mol Immunol. 18 (1), 194-205 (2021).
- Collison, L. W., Vignali, D. A. In vitro Treg suppression assays. Methods Mol Biol. 707, 21-37 (2011).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved