Method Article
Plasmid Stability Analysis with Open-Source Droplet Microfluidics
In This Article
Summary
An accessible, open-source microfluidic workflow is presented for the parallelized analysis of plasmid retention in bacteria. By employing fluorescence microscopy to quantify plasmid presence in single-cell microcolonies within gel microdroplets, this method provides a precise, accessible, and scalable alternative to traditional plate counting.
Abstract
Plasmids play a vital role in synthetic biology by enabling the introduction and expression of foreign genes in various organisms, thereby facilitating the construction of biological circuits and pathways within and between cell populations. For many applications, maintaining functional plasmids without antibiotic selection is critical. This study introduces an open-hardware-based microfluidic workflow for analyzing plasmid retention by culturing single cells in gel microdroplets and quantifying microcolonies using fluorescence microscopy. This approach allows for the parallel analysis of numerous droplets and microcolonies, providing greater statistical power compared to traditional plate counting and enabling the integration of the assay into other droplet microfluidic workflows. By using plasmids expressing fluorescent proteins alongside a non-specific fluorescent DNA stain, single colonies can be identified and differentiated based on plasmid loss or fluorescent marker expression. Notably, this advanced workflow, implemented with open-source hardware, offers precise flow control and temperature management of both the sample and the microfluidic chip. These features enhance the workflow's ease of use, robustness, and accessibility. While the study focuses on Escherichia coli as the experimental model, the method's true potential lies in its versatility. It can be adapted for various studies requiring fluorescence signal quantification from plasmids or stains, as well as for other applications. The adoption of open-source hardware broadens the potential for conducting high-throughput bioanalyses using accessible technology in diverse research settings.
Introduction
Plasmids are essential genetic elements in prokaryotic cells, significantly contributing to microbial evolution through lateral DNA transfer and rapid adaptation to environmental changes1,2. These extrachromosomal DNA molecules carry genes that provide advantageous traits, such as antibiotic resistance, metabolic functions, and virulence factors, making them valuable for microbiology research, synthetic biology, and evolution studies2. However, plasmid maintenance in cell populations is challenging due to the metabolic burden of replication and segregation, often resulting in plasmid loss without selection pressure3. Additionally, stable inheritance requires mechanisms like toxin-antitoxin and partitioning systems, adding complexity to plasmid maintenance. Assessing plasmid stability under varying conditions is crucial for both fundamental research and practical applications that utilize plasmids as a primary research element4,5. Most current methods for assessing plasmid stability have significant limitations: Flow cytometry-based methods provide indirect, population-level data, require expensive equipment, and lack direct visualization of colonies6. Bulk transcriptomics and proteomics methods are costly, provide only average cellular responses, and cannot directly quantify plasmid retention in individual colonies6. Traditional methods like serial dilution and passaging are simple but time-consuming and lack precision and representability7. Overall, quantitatively inferring or projecting the number of colonies that retain a specific functional plasmid over time or selective pressures remains challenging.
To address these challenges, a novel microfluidic workflow utilizing open hardware research instruments is presented to quantify fluorescent signals in multiple isolated colonies of bacteria, using Escherichia coli as a model. This method allows for high-throughput and precise analysis of plasmid retention over various conditions or selective pressures. Single-cell resolution analysis provides a precise method to manipulate isolated colonies, yielding sensitive data on plasmid quantification that can help to assess the retention and loss rates4.
Microfluidics, particularly droplet microfluidics, has emerged as a powerful tool for encapsulating and manipulating individual cells in controlled environments8. Specifically, microgel droplets can encapsulate single cells for high-throughput and precise analysis without the need for maintaining droplets suspended in oil9, allowing a controlled study of plasmid dynamics in a defined microenvironment. Following encapsulation of cell suspensions directly from a pipette tip10, fluorescence techniques can be used to monitor the growth of microcolonies within droplets, enabling detailed analysis of plasmid retention and segregation under different selection pressures3.
The advantages of this method over traditional bulk culture techniques include increased precision, reduced variability, and the ability to conduct high-throughput analyses. Open-source microfluidics technology overcomes the limitations of expensive proprietary systems, such as issues of accessibility, adaptability, and maintenance, which often hinder research progress11,12,13. By demonstrating how to apply the advanced experimental workflow of plasmid retention analysis in microgels with open-source instrumentation, an accessible and reliable method is provided for research in plasmid biology, synthetic biology applications, and microfluidic droplet analysis techniques.
In summary, this article presents an accessible method for quantitatively assessing plasmid retention in E. coli with high statistical power. This method's capabilities make it a valuable tool for advancing the understanding of plasmid biology and improving synthetic biology applications.
Protocol
Figure 1provides a schematic overview for assessing plasmid stability in E. coli. Details of the reagents and the equipment used are listed in the Table of Materials. The raw data and the visualization scripts are available at https://doi.org/10.17605/OSF.IO/6YWJK.
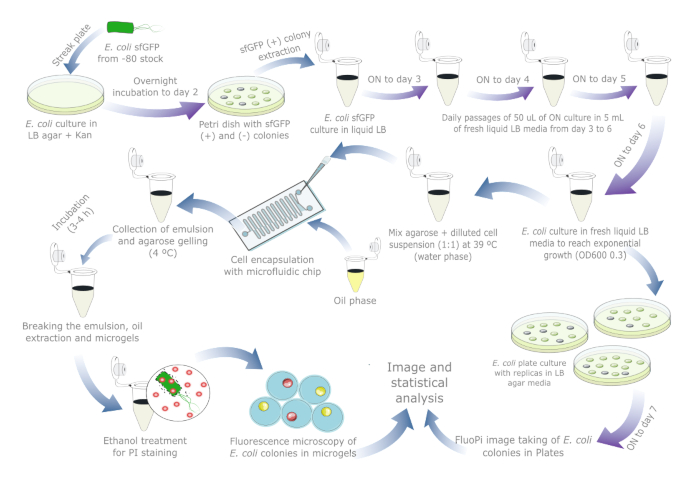
Figure 1: Day-by-day protocol for assessing plasmid stability in E. coli. Blue arrows indicate steps during the day, and purple arrows indicate overnight incubation. Every liquid and agar incubation was performed at 37 °C alongside a separate negative control tube/plate. Note that the cell culture preparation and passages are not needed for real-world samples in which plasmid loss may already have occurred, so the protocol should be reduced to one day or two if a plate-reference culture is included. Please click here to view a larger version of this figure.
1. Microfluidic chip preparation
NOTE: In this protocol, different commercial or custom chip designs can be used for cell encapsulation that is able to generate water-in-oil droplets less than 100 µm diameter in a dripping regime. For this study, the chip was designed and fabricated (see article data https://doi.org/10.17605/OSF.IO/6YWJK) following the same design and fabrication method as reported in a previously published report14.
- Obtain or prepare a PDMS-on-glass microfluidic chip using a master mold designed for gel-microdroplet generation.
- Inject a water-repellent solution (fluoroalkyl silane) into the chip to render the inner microchannels hydrophobic. Infuse the solution into the inlets and ensure all channels are filled with fluid. Leave the filled channels to rest for around 30–60 s.
- Remove the solution from the device by expelling air into the inner microchannels. Use an empty air syringe to flush the air and an absorbent wipe over the other ports to prevent splashing.
- Bake the treated device at 65 °C for 15 min on a hotplate to evaporate excess solution. Alternatively, store the device in a refrigerator (4 °C) overnight.
NOTE: The microfluidic chip is ready to use. The protocol can be paused here.
2. Sample preparation
- Cell harvesting
NOTE: Cell culture in droplets can provide relevant data on plasmid dynamics. As an experimental bacteria model, an E. coli TOP10 strain is used with the plasmid pCA_Odd1 (see deposited data https://doi.org/10.17605/OSF.IO/6YWJK) that encodes a superfolder green fluorescent protein (sfGFP) and kanamycin-resistance15. Bacteria, plasmid, and growth media can vary based on the experimental system.- Prepare Luria Bertoni (LB)-Agar by dissolving 25 g of pre-mixed LB and 12 g of agar in 800 mL of distilled water (dH2O). Also, prepare liquid LB media by dissolving 25 g of pre-mixed LB in 800 mL of dH2O. Autoclave the solutions and allow them to cool to around 60 °C. Move to a sterile environment such as a flow-hood for the next steps.
NOTE: The pre-mixed LB powder contains 10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl. Self-mixed LB-agar media can also be used. - Add 50 µL of kanamycin (prepared at 100 mg/mL) to 50 mL of liquid LB-Agar (final concentration: 100 µg/mL). Mix the solution by inverting the tube a few times.
- Pour around 15 mL of liquid LB-agar per Petri dish (90 mm x 15 mm). Prepare two plates: one for the negative control (contamination monitoring) and one for the experiment culture. Let the LB-agar solution cool and solidify until the color changes from dark to clear on both plates.
- Use a sterile loop to spread the E. coli strain on the culture plate using the streak plate technique. Place the glycerol stock E. coli back in the –80 °C immediately after use. Close both plates and incubate them overnight at 37 °C (day 1).
- On day 2, check the negative control plate for any colony as a signal of contamination (if so, repeat the steps from 2.1.4.). Identify single fluorescent colonies on the E. coli plate using a blue-light transilluminator to prepare a liquid culture stock.
- Under sterile conditions, open the plate and use a sterile loop or 200 µL pipette tip to pick the selected colony and transfer it to a culture tube with 5 mL of fresh liquid LB media and 5 µL of kanamycin (prepared at 100 mg/mL). Prepare a negative control tube without inoculation for contamination monitoring. Incubate the culture tubes at 37 °C overnight while shaking at 220 rpm.
- Passage the sample for 3 additional days in media without antibiotics to simulate conditions under which plasmid loss may occur. On days 3, 4, and 5, transfer 50 µL of the overnight culture to a new culture tube containing 5 mL of liquid LB without antibiotics. Incubate at 37 °C overnight with shaking at 220 rpm. Repeat this step until day 6, obtaining a final 5 mL culture after four total passages.
- Transfer 50 µL from the final overnight culture to a new culture tube holding 2 mL of liquid LB. Allow the culture to reach an optical density (OD600) of 0.3 using a spectrophotometer (3–4 h approximately).
NOTE: The fresh culture should be used before the OD600 increases to ensure that bacteria are in the logarithmic growth phase. If the desired concentration is not achieved, repeat from step 2.1.8.
- Prepare Luria Bertoni (LB)-Agar by dissolving 25 g of pre-mixed LB and 12 g of agar in 800 mL of distilled water (dH2O). Also, prepare liquid LB media by dissolving 25 g of pre-mixed LB in 800 mL of dH2O. Autoclave the solutions and allow them to cool to around 60 °C. Move to a sterile environment such as a flow-hood for the next steps.
- Cell-agarose mix for encapsulation
NOTE: Concentration control is essential to ensure single-cell droplet encapsulation. The required cell concentration can be calculated for a target encapsulation rate and specific droplet volume, as shown in the following example:
Droplet volume (Vdrop): If 50 µm droplets are generated
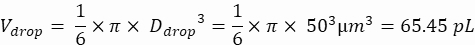
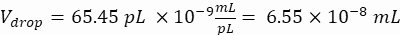
Desired cells per droplet (Cpd): One cell per five droplets on average (0.2 cells/droplet)
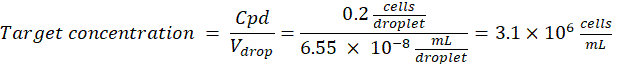
Dilution factor: The initial cell concentration is obtained in step 2.1.8
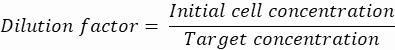
Here, 50 μm or 100 μm (without droplet splitting) droplets (65–520 pL) are generated where about one cell per five droplets is encapsulated, or 1.6 cells per droplet without splitting. For E. coli, use the conversion factor of 1 OD600 unit ≈ 7.8 x 108 cells/mL16. Multiply the OD600 value from step 2.1.8 by the conversion factor to obtain the initial concentration (cells/mL) of the culture.- Re-suspend E. coli from the prepared culture stock in liquid LB without kanamycin at a concentration of 6.2e+6 cells/mL (target concentration for 50 μm diameter droplets). Keep the bacteria suspension at room temperature until mixing with agarose.
NOTE: Antibiotics will suppress negative microcolony formation so it is essential to exclude it from the media for plasmid loss experiments. - Prepare ultra-low gelling temperature agarose by heating it to 90 °C in liquid LB at a concentration of 2% (w/v). Shake the mixture for 10 min in a temperature-controlled shaker.
- Reduce the temperature of the thermo-shaker to 39 °C to cool down the agarose solution. In parallel, place the bacteria suspension tube in the thermo-shaker for 4 min to warm it to 39 °C.
- Mix the bacteria and the agarose suspensions 1:1 ratio to obtain an agarose concentration of 1% (w/v) with cell suspension at 3.1e+6 cell/mL. Prepare the negative control solution (contamination monitoring) with the same agarose concentration using liquid LB instead of the bacteria suspension.
NOTE: The agarose-cell suspension should be used quickly to avoid concentration changes due to bacteria growth. Maintain the thermo-shaker at 39 °C to keep the agarose liquid until loaded into the tip heater for droplet generation.
- Re-suspend E. coli from the prepared culture stock in liquid LB without kanamycin at a concentration of 6.2e+6 cells/mL (target concentration for 50 μm diameter droplets). Keep the bacteria suspension at room temperature until mixing with agarose.
3. High-throughput single-colony cultivation
- Experimental setup
- Build or obtain the full open-source flow platform (see https://doi.org/10.17605/OSF.IO/6YWJK), including gas-pressure drivers and flow sensors. Alternatively, build the simpler open-source hardware strobe-enhanced microscopy stage (see build instructions https://wenzel-lab.github.io/strobe-enhanced-microscopy-stage/ and project repository https://github.com/wenzel-lab/strobe-enhanced-microscopy-stage).
NOTE: One may also use a traditional microfluidics setup with a commercial microscope, high-speed camera, and sample heating capabilities. - Integrate the open-source Raspberry Pi-based pressure and flow controller system as documented in the designated repository (https://github.com/wenzel-lab/modular-microfluidics-workstation-controller).
NOTE: The module control is illustrated and backed up to the data repository (https://doi.org/10.17605/OSF.IO/6YWJK. As an alternative, traditional pressure controllers or high-torque syringe pumps can be employed. - Include a glass slide and pipette tip heaters (https://github.com/wenzel-lab/modular-microfluidics-workstation-controller/tree/master/module-heating-and-stirring) in the setup to control the temperature of the agarose-cell sample as it enters the chip.
NOTE: These heaters, illustrated on the data repository (https://github.com/wenzel-lab/flow-microscopy-platform and https://doi.org/10.17605/OSF.IO/6YWJK), are an important feature of the open-source controller system that enables the work with agarose and may not be available in other commercial systems.
- Build or obtain the full open-source flow platform (see https://doi.org/10.17605/OSF.IO/6YWJK), including gas-pressure drivers and flow sensors. Alternatively, build the simpler open-source hardware strobe-enhanced microscopy stage (see build instructions https://wenzel-lab.github.io/strobe-enhanced-microscopy-stage/ and project repository https://github.com/wenzel-lab/strobe-enhanced-microscopy-stage).
- Single-cell encapsulation
- Position the microfluidic chip on the strobe-enhanced microscopy stage, ensuring the droplet generation junction (intersection of the aqueous and oil phases) is visible.
- Set the pipette tip heater and the glass slide heater to 40 °C using the control software interface.
- Use a syringe with tubing and PDMS plug to load the 1% agarose mix with cell suspension into a 200 μL pipette tip. Insert the tip into the tip heater and place it on the inlet of the aqueous phase in the microfluidic chip. Exchange the PDMS seal of the tip with one connected to the flow-control system tubing and start the infusion of the cell suspension.
- Insert the outlet tubing end into a waste tube and set flow rates or pressures of two phases on the user interface to deliver fluid slowly to the microfluidic channel. Use 200 µL/h (180 mbar) for the aqueous phase and 1700 µL/h (320 mbar) for the oil phase. Allow 1 min for the stabilization of droplet generation.
NOTE: Pressure values depend on the channel sizes of the chip design, and flow values may have to be adjusted for different droplet generation junction designs. - Once the droplet generation is stable, transfer the waste and collection tubing to the collection tube. Continuing collecting droplets until the sample reservoir is empty. Repeat steps 3.2.3 to 3.2.5 to encapsulate the negative control solution from step 2.2.4.
NOTE: Sample collection should be completed within 15 min. - Store the collection tubes on ice during the droplet generation or place them at 4 °C after the experiment for 1 h to allow agarose to gel inside the droplets.
NOTE: The microfluidic chip can be reused if the microchannels remain unclogged and the same suspension is loaded. Discard the pipette tip after completing emulsion generation (steps 3.2.3–3.2.5).
- Colony growth and release from emulsion
- Transfer the gel microdroplets containing bacteria and the negative control in droplets to an incubation chamber set to 37 °C.
- Incubate the microdroplets for at least 4 h, or overnight, to allow for sufficient colony growth. Confirm that the negative control shows no signs of contamination via bright field microscopy.
- To release the colonies from the emulsion, remove as much oil as possible from below the gel microdroplet emulsion using a pipette or a syringe with a needle (here, size 21 G needle was used).
- Transfer 50 µL of the gel microdroplets to a new microtube and store it at 4 °C for further droplet analysis. To the remaining emulsion, add a 1:1 mixture of fluorinated oil with 1H,1H,2H,2H-perfluoro-1-octanol (PFO) in a volume equal to the emulsion.
- Add approximately 200 µL of phosphate-buffered saline (PBS) buffer, or 0.9% w/v NaCl buffer, on top of the emulsion. Vortex the mixture and briefly spin it down in a fixed-speed centrifuge.
- Carefully remove the oil phase from the bottom of the liquid interface and discard 100 µL of PBS from the top. Repeat steps 3.3.4–3.3.5 to obtain washed microgels in PBS buffer with minimal or no oil residue.
NOTE: The microgels settle at the liquid interface; avoid removing them along with the oil phase.
4. Single-colony analysis
- Cell staining
NOTE: Many different stain combinations work for this protocol. In essence, a DNA or cell-wall stain that has a color distinct from the fluorescent protein encoded by the plasmid should be chosen and that can be analyzed using the available filter combinations on the fluorescence microscope. Here, the DNA of cells was stained with Propidium Iodide (PI) to distinguish its fluorescence from the green fluorescent protein encoded on the plasmids, but many other DNA stains can be used.
CAUTION: PI is a potential carcinogen and must be handled with appropriate personal protective equipment. Dispose of the dye safely and in compliance with local regulations.- Centrifuge the washed microgels at approximately 80 x g for 5 min at room temperature. Discard the supernatant using a pipette.
- Transfer 50 uL of the microgels to a new microtube to serve as a negative control for ethanol treatment.
NOTE: PI stain only enters cells with compromised membranes, such as those affected by ethanol treatment. - Add an equal volume of 70% ethanol to the remaining microgels and mix briefly with a vortex. Incubate at room temperature for 15 min to permeabilize bacterial membranes for PI staining. Repeat step 4.1.1.
- Add an equal volume of 0.9% w/v NaCl to the microgels and briefly vortex. Repeat step 4.1.1.
- Add 2 µL of PI (1 mg/mL) to both microtube samples. Mix thoroughly and incubate in the dark at room temperature for 15 min.
NOTE: If the negative control presents any red fluorescent signal via fluorescence microscopy, the integrity of colonies may have been compromised during their exposure to other solutions in previous steps.
- Microscopy
NOTE: Droplets and microgels are imaged with an inverted epi-fluorescence microscope to obtain the droplet size distribution and the fluorescence of bacterial colonies in microgels (Figure 2). Here, an open-source inverted microscopy platform (https://github.com/wenzel-lab/SQUID-bioimaging-platform)17 is used with a 10x 0.3NA objective, a white LED array for brightfield illumination, and a 470nm LED for excitation. Commercial epi-fluorescence microscopes can be used to image droplets and microgels. Calibration is required because illumination and filters vary depending on the model, brand, and fluorescent proteins used.- Transfer 2 μL of gel microdroplets into an imaging chamber slide and add 5 μL of fluorinated oil to help form a monolayer of droplets for optimal imaging.
NOTE: Cell counting chambers or simple microfluidic chambers can help spread the emulsion thinly and slow the drying process. - On the microscope, activate the white LED-matrix illumination from the top for brightfield imaging. Mount the prepared slide, focus on the sample, and locate a monolayer of droplets. Capture a brightfield image.
- Without moving the sample, capture a fluorescence image of the colonies by switching to the 470 nm LED for excitation. Adjust the filter wheel to align with the green wavelength filter for sfGFP imaging. Scan all the areas containing monolayers of droplets and repeat steps 4.2.2–4.2.3 to ensure statistical robustness on droplet analysis.
- Transfer 2 µL of the stained microgels into an imaging chamber chip and 5 µL of 0.9% w/v NaCl to help form a monolayer of microgels. Seal the inlet and outlet of the chip to prevent evaporation during imaging.
NOTE: Evaporation can affect the colocalization of the fluorescent labels. Microfluidic chambers are recommended for prolonged imaging. - Repeat steps 4.2.2–4.2.3. Adjust the red wavelength interval filter for PI imaging and capture the respective images.
- After completing imaging at one location, find the next suitable area on the slide with a monolayer of microgels and repeat the imaging process to ensure statistical robustness and comprehensive analysis of the sample.
- Transfer 2 μL of gel microdroplets into an imaging chamber slide and add 5 μL of fluorinated oil to help form a monolayer of droplets for optimal imaging.
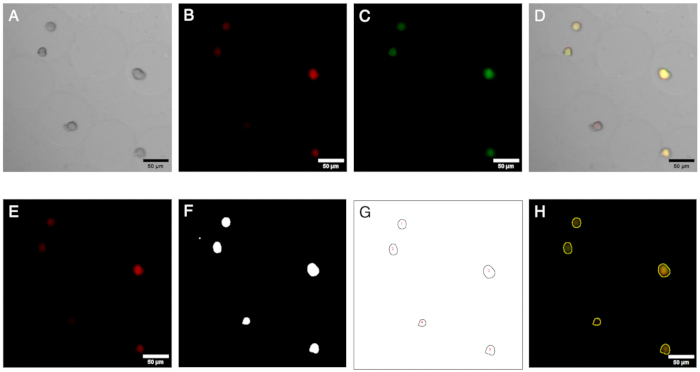
Figure 2: Microscopy images and their analysis. Fluorescence imaging and analysis of colonies in microgels. (A–C) Image channels acquired via brightfield and fluorescence microscopy with the inverted microscope. The composite image (D) shows the presence of a negative colony (red fluorescence only) within the microgels. (E–H) Outcomes of the image analysis workflow. By generating ROIs, colonies can be identified on red and green channels, and the signals can be quantified to define the presence of negative colonies. Scale bars: 50 µm. Please click here to view a larger version of this figure.
- Image analysis
NOTE: To analyze the fluorescence signals from the encapsulated colonies and identify rare events, images from brightfield and fluorescence imaging can be processed with Fiji/ImageJ (Figure 3). These steps can be implemented into a macro script, and the parameter values may vary depending on the optical configuration.- Open the images from the green and red channels. Define a rectangular region of interest (ROI) starting at coordinates (500, 500) with a width and height of 2000 pixels each.
NOTE: These parameters apply to 3000 x 3000 pixel images, and the ROI defines the area with better illumination. - Crop the image to the defined rectangular ROI. Subtract a constant value of 10 from each pixel's intensity and apply a smoothing filter to the image to reduce background noise and make the objects of interest more distinct.
NOTE: Only this region will be used for further analysis. - Duplicate the image from the red channel and convert it into a binary mask to identify the rare events related to the plasmid. Set the threshold values between 10 and 255. Pixels within this range will be considered foreground (objects of interest), while others will be treated as background.
- Perform morphological operations to close small gaps and fill any holes within the objects of interest. Apply the watershed algorithm to separate overlapping objects within the binary mask.
- Analyze the particles in the binary mask. Only consider particles with a size greater than 30 pixels and a circularity between 0.50 and 1.00. The results are summarized and added to the results table. Save the set of ROIs detected by the particle analysis for further visualization.
- Display the saved ROIs on the images from the green and red channels. Measure the intensity or other properties of the ROIs within these images. Record the measurements in the results table and save the results in separate CSV files for further statistical analysis.
- Open the images from the green and red channels. Define a rectangular region of interest (ROI) starting at coordinates (500, 500) with a width and height of 2000 pixels each.
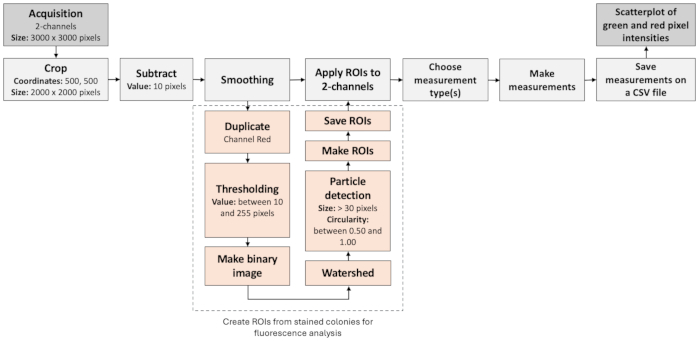
Figure 3: Image analysis workflow to identify negative colonies. The figure illustrates a step-by-step workflow to automatically process and assess fluorescence images. The workflow is based on the colocalization of fluorescence labels as well as particle analysis. Please click here to view a larger version of this figure.
5. Agar plate comparison assay
NOTE: To compare the droplet method to a traditional plate assay, the quantification of fluorescent colonies of the same E. coli strain was obtained in step 2.1.8. was performed using Petri dishes. This served as an analog control method to measure sfGFP plasmid stability. See also method illustration in Figure 1.
- Prepare LB-agar (without antibiotic) as described in step 2.1.1 from the Cell Harvesting section. Cool the agar below 60 °C and homogenize.
- In a sterile environment, pour 15 mL of the agar medium into each Petri dish. Allow the plates to solidify with lids partially open until they are ready for use.
- Inoculate three plates with 10 µL each of an E. coli culture in LB media with an OD600 of 0.007 ± 0.002. Spread evenly using an L-shaped spreader. Incubate the plates at 37 °C, closed and inverted. Prepare a control plate without bacteria under the same conditions.
- After 24 h, capture fluorescence images of the plates using the FluoPi, an open-source fluorescence imaging system (https://github.com/RudgeLab/FluoPi)15.
NOTE: The FluoPi consists of a Raspberry Pi camera with a blue excitation light at a wavelength centered at 470 nm and acrylic excitation and emission filters. - Manually count the fluorescent and non-fluorescent colonies in each plate using the captured images.
Results
Validation of cell encapsulation and microcolony formation
The cell encapsulation can be visually confirmed by performing brightfield microscopy on the gel microdroplets before breaking the emulsion and washing the microgels. A representative result of the emulsion at this step is shown in Figure 4.
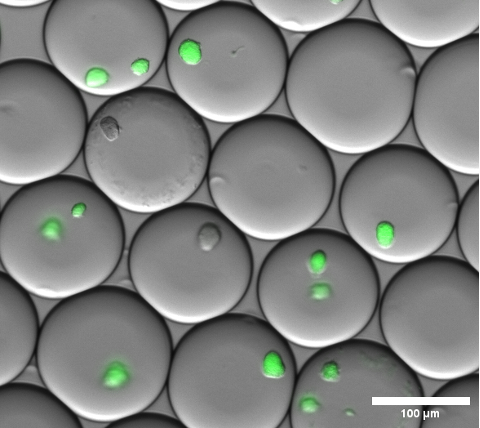
Figure 4: Section of a fluorescence microscopy overlay image. After overnight incubation, representative microcolonies of sfGFP expressing E. coli colonies inside of gel microdroplets. A microscope objective with 10x magnification and a 0.30 NA was used. Scale bar: 100 µm. Please click here to view a larger version of this figure.
Image analysis outcomes
Once microgels are stained and brightfield, as well as fluorescence channels acquired at several positions, the colonies identified as negative in the original images can be visualized (see Figure 5A). Data extracted from all images of one experiment can be plotted to show the fluorescence ratio of various colonies, highlighting those that lost plasmid-encoded fluorescence (see Figure 5B). Results indicate that 100 colonies lost their plasmid or plasmid functionality from a total of 2785 analyzed microcolonies, corresponding to 3.6%.
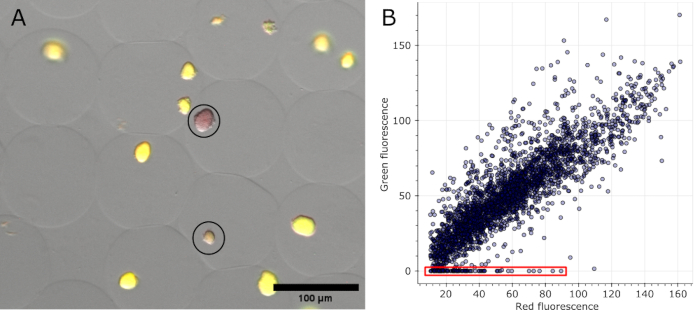
Figure 5: Quantification of negative microcolonies. (A) Section of a fluorescence microscopy overlay image. After oil removal and staining, representative colonies in microgels expressing sfGFP and two negative colonies showed the red fluorescence of the DNA stain (circled in black). Scale bar: 100 µm. (B) Scatterplot of the fluorescence values of individual microcolonies extracted from 16 multi-channel microscopy images. Colonies without any green fluorescence were counted as negative, as indicated in red in the graph. Please click here to view a larger version of this figure.
Agar plate quantification
Images of the triplicate plates are shown in Figure 6, with non-fluorescent colonies indicated by white arrows. The first plate (Figure 6A) displayed a total of 213 colonies, of which 1 was not fluorescent. The second plate (Figure 6B) had a total of 49 colonies, with no non-fluorescent colonies. The third plate (Figure 6C) showed a total of 252 colonies, 6 of which were not fluorescent. These results correspond to a mean colony plasmid loss rate of 2.3%, with a large standard deviation of 3.2.
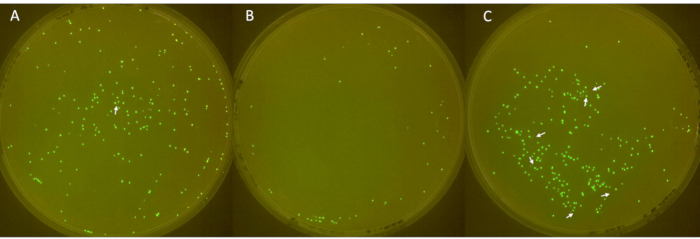
Figure 6: Identification of negative colonies on plates. (A–C) Fluorescent and non-fluorescent E. coli colonies on LB-agar plates (diameter: 90 mm, height: 15 mm). The inoculum, derived from E. coli with sfGFP from -80 °C stock, was streaked on day 1, cultured with antibiotic on day 2, and diluted 1:100 daily from day 3 to 6 to allow plasmid loss. Colonies were incubated at 37 °C for 24 h and imaged in a FluoPi chamber. Non-fluorescent colonies were enhanced with GIMP and indicated with white arrows. Please click here to view a larger version of this figure.
Discussion
A gel microdroplet-based method is demonstrated to effectively identify and quantify colonies with and without plasmid-encoded genetic expression of fluorescent proteins, such as sfGFP. Colonies that do not sufficiently express the plasmid product are identified using a fluorescent DNA stain (here, Propidium Iodide) that stains all colonies and features a different emission wavelength. This integration of droplet microfluidics, gelling, and fluorescence microscopy, utilizing open-source technology, enables executing an advanced workflow in many research settings11,13. The successful generation of gel-microdroplets enables advanced molecular biology single-cell workflows, including cell lysis, single genome amplification, metabolic cell interaction screens, media exchange, and more8,9. These advantages are used in this protocol to grow, stain, and analyze microcolonies in a more scalable fashion than in traditional plate-based assays.
Critical steps
The encapsulation process is a critical and delicate part of the protocol. Precise control of ingredient concentrations, flow rates, and pressures is required to generate uniform microgels within a specific size range and control the average number of cells per droplet. Furthermore, maintaining the concentration and temperature of the cell-agarose mix prevents clumping or premature gelation. The temperature control of the liquid agarose-cell suspension in a pipette tip is a particularly advantageous implementation of our open-source hardware microfluidics workstation that provides much easier and more robust microgel generation compared with efforts to control the temperature of syringe pumps and tubing. Since cells are mixed with the agarose growth medium before encapsulation and cultivation, the agarose microgels have to be generated quickly in order to avoid major cell concentration changes. For this purpose, a droplet-splitting microfluidic chip design inspired by Abate et al. was optimized18.
Modifications and troubleshooting
Several calibrations and modifications were necessary to refine the original protocol. The encapsulation of agarose is much more challenging than regular water-in-oil droplets, requiring the design of a system to maintain the agarose in a liquid state while ensuring the aqueous phase flow achieves a homogeneous particle size range. Changes in agarose viscosity due to gelation affect the flow rate, leading to larger particle sizes. The microscopy requires a careful selection of filters and light sources to ensure non-overlapping excitation and emission signals for clear differentiation. Initially, DAPI was chosen for staining bacteria, but its emission signal overlapped with sfGFP, causing sfGFP to be detected in the blue detection channel. We switched to PI because its emission is well-separated from sfGFP at long wavelengths (red light).
While plasmid loss was quantified using the proposed method, the sfGFP plasmid used was unexpectedly stable, displaying hardly any instances of plasmid loss in the first generation of cells cultivated without antibiotics, even under stress conditions such as pH9 media and incubation at 40 °C. This observation is consistent with the findings of other research groups1,19. The plasmid stability limited the demonstration of the method's full quantification capabilities for initial cell culture generations, but it did demonstrate that the method is sensitive enough to detect even small differences in plasmid retention. The observation of high-plasmid stability in early generations has an important implication for droplet microfluidic screens using a negative selection assay, such as target bacteria inhibition. It means that the plasmid loss of selection targets is a low source of false-positive selection results. As droplet microfluidic screens typically exceed other high-throughput screens, such as pipetting robot workflows, by orders of magnitudes in throughput, these rare events need to be assessed and taken into account.
Limitations
Despite its advantages, there are limitations to the presented method. Microfluidic device fabrication requires expertise and meticulous attention to detail, as well as tight experimental control of flow rates to ensure deterministic encapsulation efficiency. These aspects may require optimization for different experimental setups. While this method relies on fluorescence microscopy for signal detection, necessitating access to suitable imaging equipment, this equipment can be fabricated using open-source hardware, making it more accessible. Furthermore, microgels can be processed in commercial flow cytometry with large nozzles, further improving accessibility and experimental throughput. Droplet sorters can also be used for this cytometric analysis.
Moreover, while the method is designed to detect fluorescent signals from plasmids, stains, or other markers, it is limited to cells that can be fluorescently labeled, which may not apply to all bacterial strains or experimental conditions. However, the method can be adapted to incorporate other types of microscopies, such as phase-contrast or brightfield microscopy, allowing for phenotyping applications beyond fluorescence. Additionally, it can be combined with spectroscopic techniques like FTIR or Raman spectroscopy, expanding its capabilities to analyze chemical compositions and structural information of the encapsulated cells. These adaptations broaden the range of its applicability, making it a versatile tool for diverse research settings.
Significance and applications
Traditional assays for plasmid loss19 do not allow a good quantification of the ratio of cells that lost their expression, information that can be very important in experimental method design and various biological applications. Usually, colony types are enumerated in agar plate assays, where well-defined isolated colonies can be obtained, as demonstrated in Figure 4. However, overlapping colonies are challenging to identify with confidence; in our hands, we do not always obtain an optimal colony density, and many plates are necessary to obtain good statistics of low-frequency plasmid loss events. The proposed method offers a more robust approach to accurately quantify fluorescent signals coming from isolated colonies with a higher number of colonies than the agar plate analog methods because, in microdroplets, colonies develop separately, are smaller, and are easy to load into imaging chambers, enabling microscopy or flow-cytometry based quantification of large colony numbers. This can significantly improve the statistical representation of the method and allow integration into other gel-microdroplet workflows.
The usage of open-source hardware11,20 allows researchers to customize the microfluidic workstation design and precisely adjust flow rat; therefore, particle size supports various cell types and experimental conditions. This flexibility extends to potentially incorporating other microscopy types, such as phase-contrast or spectroscopy, broadening the method's applicability. The method's capacity to evaluate plasmid stability under various conditions is crucial for applications requiring plasmid retention without antibiotic selection, under particular stress conditions, or various culture generations. The versatility and adaptability of the presented method make it valuable for diverse research applications in fields including synthetic biology, environmental monitoring, and clinical diagnostics2.
Disclosures
The authors declare that no competing financial interests or personal relationships could have influenced the work reported in this paper.
Acknowledgements
This work is part of funded projects granted to T.W. from ANID FONDECYT Regular 1241621 and the Chang Zuckerberg Initiative project ‘Latin American Hub for Bioimaging Through Open Hardware’. T.W. is also grateful for funding from CIFAR, as Azrieli Global Scholar in the CIFAR MacMillan Multiscale Human program.
Materials
| Name | Company | Catalog Number | Comments |
| 1H,1H,2H,2H-Perfluoro-1-octanol | Sigma-Aldrich | 370533-25G | For breaking emulsion |
| 70% ethanol | For cell permeabilization | ||
| Agar-Agar | Winkler | 9002-18-0 | |
| Biopsy Punch | 0.75 mm and 1.8 mm | ||
| Blue LED transilluminator | IO Rodeo | ||
| Culture tube | 15 mL | ||
| Desiccator | With vacuum pump | ||
| Disposable cup | For mixing PDMS | ||
| Disposable fork | For mixing PDMS | ||
| E. coli TOP10 strain | |||
| FluoPi microscope | https://github.com/wenzel-lab/FluoPi | Green fluorescence imaging system for analyzing plates | |
| Fluorinated Oil | 3M | Novec 7500 | |
| Glass slide heater | https://github.com/wenzel-lab/modular-microfluidics-workstation-controller/tree/master/module-heating-and-stirring | For controlling the temperature at 40 °C of microfluidic chip | |
| Glass Slides | |||
| Hotplate | Mechanic | For evaporating Aquapel | |
| Image analysis software | Fiji/ImageJ | 2.14.0/1.54f | |
| Incubator | Mundo Lab | MLAB Scientific / For incubation of plates and microgels | |
| Isopropanol | For cleaning glass slides | ||
| Kanamycin | 100 ug/mL concentration | ||
| L-shaped spreader | For spreading bacteria on agar plates | ||
| Master mold | Chip design on silicone or glass wafer | ||
| Microtubes | 2 mL | ||
| NaCl solution | Sodium chloride 0.9% w/v | ||
| Open-source hardware strobe-enhanced microscopy stage | https://github.com/wenzel-lab/flow-microscopy-platform | For bright-field microscopy | |
| Petri dish | Citotest | 2303-1090 | 90 x 15 mm |
| Pipette tip heater | https://github.com/wenzel-lab/modular-microfluidics-workstation-controller/tree/master/module-heating-and-stirring | For controlling the temperature at 40 °C of pipette tip | |
| Plasma Cleaner | Diener Electronic | 117056 | For bonding PDMS with a glass slide |
| Plasmid pCA_Odd1 | Encodes sfGFP and kanamycin resistance | ||
| Polytetrafluoroethylene (PTFE) tubing | Adtech Polymer Engineering Ltd | ||
| Pre-mixed Luria Bertoni medium | US Biological Life Science | L1520 | |
| Propidium iodide (PI) | For staining | ||
| Raspberry Pi-based pressure and flow controller system | https://github.com/wenzel-lab/modular-microfluidics-workstation-controller | For controlling pressure and flow rates | |
| Silicone elastomer base | Sylgard | PDMS kit - 184 Silicone Elastomer Kit | |
| Silicone elastomer curing agent | Sylgard | PDMS kit - 184 Silicone Elastomer Kit | |
| Spectrophotometer | For measuring absorbance | ||
| SQUID microscope | https://github.com/wenzel-lab/SQUID-bioimaging-platform | Multi-fluorescence imaging system for analyzing stained cells | |
| Sterile loop | For picking a colony and streaking plating | ||
| Surfactant | Sphere Fluidics | Pico-Surf | |
| Syringes | NIPRO | With filters and tubing | |
| Temperature-controlled shaker | Mundo Lab | DLAB HCM100-Pro | |
| Tweezer | |||
| Ultra-low gelling temperature agarose | Sigma-Aldrich | A2576-5G | For generating hydrogel beads |
| Water repelent solution (fluoroalkyl silane) | Aquapel | For treating microchannels of PDMS device |
References
- Wein, T., Hülter, N. F., Mizrahi, I., Dagan, T. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat Commun. 10 (1), 2595(2019).
- Rodríguez-Beltrán, J., DelaFuente, J., León-Sampedro, R., MacLean, R. C., Millán, ÁS. Beyond horizontal gene transfer: The role of plasmids in bacterial evolution. Nat Rev Microbiol. 19 (6), 347(2021).
- Wein, T., Dagan, T. Plasmid evolution. Curr Biol. 30 (19), R1158-R1163 (2020).
- Chen, S., Larsson, M., Robinson, R. C., Chen, S. L. Direct and convenient measurement of plasmid stability in lab and clinical isolates of E. coli. Sci Rep. 7 (1), 4788(2017).
- Rouches, M. V., Xu, Y., Cortes, L. B. G., Lambert, G. A plasmid system with tunable copy number. Nat Commun. 13 (1), 3908(2022).
- Silva, F., Queiroz, J. A., Domingues, F. C. Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol Adv. 30 (3), 691-708 (2012).
- Wang, R., et al. Construction of novel pJRD215-derived plasmids using chloramphenicol acetyltransferase (cat) gene as a selection marker for Acidithiobacillus caldus. PLoS ONE. 12 (8), e0183307(2017).
- Moragues, T., et al. Droplet-based microfluidics. Nat Rev Methods Primers. 3 (1), 32(2023).
- Vitalis, C., Wenzel, T. Leveraging interactions in microfluidic droplets for enhanced biotechnology screens. Current Opinion in Biotechnology. 82, 102966(2023).
- Sinha, N., Subedi, N., Wimmers, F., Soennichsen, M., Tel, J. A pipette-tip based method for seeding cells to droplet microfluidic platforms. J Vis Exp. (144), e57848(2019).
- Wenzel, T. Open hardware: From DIY trend to global transformation in access to laboratory equipment. PLOS Biol. 21 (1), e3001931(2023).
- Murillo, L. F. R., Wenzel, T. Welcome to the journal of open hardware. J Open Hardware. 1 (1), (2017).
- Shin, J. H., Choi, S. Open-source and do-it-yourself microfluidics. Sens Actuators B Chem. 347, 130624(2021).
- Pryszlak, A., et al. Enrichment of gut microbiome strains for cultivation-free genome sequencing using droplet microfluidics. Cell Rep Methods. 2 (1), 100137(2021).
- Pollak, B., et al. Universal loop assembly: open, efficient and cross-kingdom DNA fabrication. Biology. 5 (1), (2020).
- Volkmer, B., Heinemann, M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS ONE. 6 (7), e23126(2011).
- Li, H., et al. Squid: Simplifying quantitative imaging platform development and deployment. bioRxiv. , (2020).
- Abate, A. R., Weitz, D. A. Faster multiple emulsification with drop splitting. Lab Chip. 11 (11), 1911-1915 (2011).
- Lau, B. T. C., Malkus, P., Paulsson, J. New quantitative methods for measuring plasmid loss rates reveal unexpected stability. Plasmid. 70 (3), 353-361 (2013).
- Oellermann, M., et al. Open hardware in science: The benefits of open electronics. Integr Comp Biol. 62 (4), 1061-1075 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved