Method Article
Diagnóstico postmortem de la rabia en animales mediante el ensayo de reacción en cadena de la polimerasa con transcriptasa inversa LN34 en tiempo real multiplexado actualizado
En este artículo
Resumen
Este protocolo demuestra el ensayo de reacción en cadena de la polimerasa con transcriptasa inversa (RT-PCR) pan-lyssavirus LN34 en tiempo real desde la recolección de tejidos hasta la interpretación de los resultados, incluidas las actualizaciones de las secuencias de cebadores y formulaciones para mejorar el rendimiento del ensayo para algunos lyssavirus y lagomorfos no antirrábicos. También demostramos la configuración del ensayo para un formato multiplexado LN34 de un solo pocillo (LN34M).
Resumen
La rabia es una enfermedad zoonótica mortal causada por la rabia por Lyssavirus (RABV) y los virus de ARN hebra negativa relacionados del género Lyssavirus (familia Rhabdoviridae). El ensayo LN34 se dirige a la región líder altamente conservada y al gen de nucleoproteína del genoma del lyssavirus y utiliza cebadores degenerados y una sonda TaqMan que contiene nucleótidos bloqueados para detectar ARN en los diversos géneros de Lyssavirus . Un resultado negativo para la rabia debe hacerse solo si se examina una sección transversal completa del tronco encefálico y tres lóbulos del cerebelo; sin embargo, la identificación del ARN de lyssavirus en cualquier tejido es diagnóstica de infección por rabia. El tejido se recoge y homogeneiza en el reactivo TRIzol, que también inactiva el virus. La extracción de ARN se realiza utilizando un kit de extracción comercial basado en columna de centrifugación. Las mezclas maestras se preparan en un espacio limpio y se alícuota en una placa de 96 pocillos antes de agregar la muestra de ARN. En entornos clínicos, cada muestra se analiza mediante RT-PCR en tiempo real para detectar la presencia de ARN de lyssavirus por triplicado y por separado para el ARNm de β actina del huésped. Los controles positivos y negativos se incluyen en los pasos de extracción y RT-PCR en tiempo real del protocolo. El análisis de datos implica el ajuste manual de los umbrales para estandarizar los valores de Ct en todas las ejecuciones de instrumentos. Los resultados positivos están determinados por la presencia de amplificación típica en el ensayo pan-lyssavirus (Ct ≤ 35). Los resultados negativos están determinados por la ausencia de amplificación típica en el ensayo pan-lyssavirus y la detección de ARNm de β-actina del huésped (Ct ≤ 33). La observación de valores fuera de estos rangos o la falla de los controles de ensayo pueden invalidar la ejecución o dar lugar a resultados no concluyentes para una muestra. El protocolo debe seguirse de cerca para garantizar una alta sensibilidad y especificidad del ensayo. Las modificaciones del procedimiento pueden afectar al rendimiento del ensayo y dar lugar a resultados falsos positivos, falsos negativos o no interpretables.
Introducción
Este protocolo describe el procedimiento para las pruebas diagnósticas de rabia utilizando el ensayo de reacción en cadena de la polimerasa con transcriptasa inversa (RT-PCR) en tiempo real del pan-lyssavirus LN34 desde la recolección de la muestra hasta la interpretación de los resultados. El procedimiento se dividirá en tres secciones: recolección de muestras cerebrales según sea relevante para el ensayo LN34 (Sección 1), extracción manual de ARN basada en columna utilizando el kit Direct-zol RNA Miniprep (Zymo Research R2051) (Sección 2) y configuración del ensayo RT-PCR en tiempo real LN34 utilizando el kit AgPath-ID RT-PCR de un solo paso (ThermoFisher Scientific AM1005) (Sección 3). La extracción de ARN y la RT-PCR se pueden realizar con otros productos, pero los kits deben validarse antes de su uso para garantizar que el ARN de Lyssavirus se extraiga y amplifique adecuadamente.
En la sección 1 se describe la recogida de los tejidos cerebrales adecuados que se utilizarán en el ensayo RT-PCR en tiempo real LN34. No se incluye la descripción de la necropsia del animal, la decapitación y la extracción del cerebro. Las muestras pueden contener agentes infecciosos. Se deben seguir los procedimientos de bioseguridad detallados en la Bioseguridad en Laboratorios Microbiológicos y Biomédicos 6ª Edición1 para mitigar el riesgo. Las muestras deben considerarse infecciosas hasta que se complete la inactivación. La inactivación viral y la validación de los ensayos deben realizarse en cada laboratorio de acuerdo con los estándares de esa institución. Los laboratorios deben seguir los procedimientos estándar de seguridad y calidad determinados por su institución al implementar una nueva prueba diagnóstica.
Con base en lo que se sabe sobre la propagación del virus de la rabia durante la infección, el tronco encefálico y el cerebelo son los mejores tejidos para el diagnóstico de la rabia, y estos tejidos son recomendados para las pruebas de rabia por la Organización Mundial de la Salud y la Organización Mundial de Sanidad Animal 2,3,4,5. Debido a que la diseminación del virus puede ser unilateral (Figura 1), especialmente en animales más grandes, se debe examinar una sección transversal completa del tronco encefálico y tres lóbulos del cerebelo para descartar la rabia. En el caso de las muestras que no cumplan con estos criterios mínimos, el laboratorio puede rechazar la muestra por considerarla insuficiente para el análisis u optar por realizar el análisis con fines de vigilancia o de regla. Si no se reciben los tejidos requeridos, pero el laboratorio opta por analizar la muestra, un resultado negativo de la prueba debe interpretarse como no concluyente para la rabia para ese animal porque la presencia de ARN viral en otros tejidos puede ser tardía, de baja abundancia, intermitente o inexistente. Es necesario recolectar las muestras requeridas o realizar pruebas adicionales para descartar la rabia en ese caso. Sin embargo, la identificación del ARN del lyssavirus en cualquier tejido es diagnóstica de infección por rabia 3,6. Ejemplos de muestras que se pueden analizar para detectar el ARN del virus de la rabia para la infección por rabia (pero no para descartarla) son la corteza, el hipocampo, la médula espinal, las muestras degradadas, la piel, la saliva y la córnea. Se debe realizar una evaluación cualitativa del estado de cada muestra a su llegada al laboratorio. La refrigeración puede conservar una muestra durante al menos 72 horas, pero no debe usarse a largo plazo. Los ciclos repetidos de congelación y descongelación pueden reducir la sensibilidad de la prueba, y se deben evitar más de cinco ciclos de congelación y descongelación. Si el estado del tejido impide la identificación segura de las estructuras cerebrales, la muestra debe identificarse como insatisfactoria. En el caso de un espécimen insatisfactorio, aún se pueden realizar pruebas para descartar (pero no descartar) la rabia. Los resultados positivos de las pruebas se informan como tales. Los resultados negativos o no concluyentes en el tejido insatisfactorio deben informarse como no concluyentes para evitar interpretaciones erróneas como un diagnóstico negativo.
Este protocolo se desarrolló a partir de los procedimientos publicados 7,8,9 e incluye cebadores actualizados dirigidos a la región líder del genoma del lyssavirus y a la secuencia codificante de nucleoproteínas. La sonda se dirige a una secuencia corta y altamente conservada y utiliza nucleótidos bloqueados para permitir una detección amplia. El ensayo detecta ARN de diversos lisavirus a diferentes concentraciones8. Este protocolo demuestra los procedimientos de laboratorio para realizar el ensayo de PCR en tiempo real LN34, pero la detección precisa y sensible del ARN del lisavirus depende de otros elementos que no están ampliamente cubiertos en este protocolo, como el almacenamiento de muestras, el mantenimiento de registros, la capacitación/competencia del personal, el seguimiento de resultados, la interpretación de resultados, el aseguramiento de la calidad, las medidas de seguridad del laboratorio y la resolución de problemas. Los ensayos basados en PCR son propensos a la contaminación cruzada debido a su alta sensibilidad. La contaminación cruzada puede evitarse siguiendo las buenas prácticas de laboratorio, como cambiar con frecuencia los guantes, manipular una muestra a la vez, desinfectar las superficies de trabajo con agentes descontaminantes eficaces entre muestras y mantener los tubos cerrados y las muestras separadas de los reactivos de PCR. Los reactivos y las muestras de PCR se pueden separar fácilmente empleando un flujo de trabajo unilateral y separando las áreas de trabajo de preamplificación y posamplificación. Por ejemplo, prepare mezclas maestras de PCR en un lugar físicamente separado de donde se manipulan las muestras. Cambie los guantes con frecuencia para evitar la contaminación de los reactivos de PCR con muestras, residuos o ARN de control positivo. La placa o los tubos de PCR deben moverse después de la adición de la mezcla maestra a una segunda ubicación donde se pueda agregar la muestra y el ARN de control. Es importante destacar que los productos de PCR no deben manipularse en áreas donde se preparan muestras o mezclas maestras.
No hay sustituto para la práctica y la experiencia al realizar pruebas diagnósticas. Todos los nuevos empleados deben ser capacitados, y el personal de pruebas debe ser evaluado para determinar su competencia al menos una vez al año, siguiendo los requisitos del director de laboratorio correspondiente. Cualquier observación de resultados inusuales o fallas en el ensayo debe anotarse, investigarse y corregirse de inmediato. Cada nuevo lote de reactivos debe validarse utilizando muestras con valores de Ct conocidos (como un control positivo o una muestra archivada). Todo el equipo debe someterse a un mantenimiento de rutina, según lo sugerido por el fabricante, y el rendimiento del ensayo debe verificarse después de cualquier mantenimiento o reparación. Los niveles de temperatura deben controlarse en el equipo correspondiente para garantizar que los refrigeradores y congeladores se mantengan dentro de los criterios establecidos para un rango de temperatura aceptable para los reactivos utilizados en las pruebas de diagnóstico.
Las modificaciones del procedimiento pueden afectar al rendimiento del ensayo y pueden dar lugar a falsos positivos, falsos negativos o resultados no interpretables. Las recomendaciones deben seguirse de cerca para garantizar una alta sensibilidad y especificidad del ensayo. Un laboratorio que desee incorporar modificaciones a este protocolo debe validar y confirmar los métodos modificados en consulta con los CDC.
Protocolo
Las muestras de tejido cerebral postmortem se obtuvieron a través de actividades rutinarias de vigilancia o diagnóstico de la División de Poxvirus y Rabia (CDC; Atlanta, Georgia, Estados Unidos).
1. Recolección de tejido cerebral para el diagnóstico postmortem de rabia en animales mediante el ensayo RT-PCR en tiempo real LN34 pan-lyssavirus
NOTA: Las muestras pueden contener agentes infecciosos. Use equipo de protección personal (EPP) adecuado (guantes de goma gruesos u otros guantes resistentes a los cortes, bata de laboratorio, delantal impermeable, mascarilla quirúrgica, botas, fundas protectoras y un protector facial) y siga las normas de seguridad requeridas para el uso, almacenamiento y eliminación de muestras. Se requiere la vacunación antirrábica previa a la exposición, pruebas serológicas periódicas e inmunizaciones de refuerzo (según sea necesario) para cualquier persona antes de trabajar, probar, producir o realizar actividades de investigación con lyssavirus o especímenes conocidos o potencialmente infectados 2,3,4,6,10.
- Etiquete un tubo de recolección de muestras por muestra con una etiqueta de acceso. Llene cada tubo de recogida de muestras con 1 ml de reactivo TRIzol u otro tampón de homogeneización y una porción de perlas MagNA Lyser (en adelante, "perlas cerámicas"). Para agregar cuentas de cerámica, vierta con cuidado desde el tubo de cuentas en el tubo de recolección de muestras. Los tubos de perlas de cerámica generalmente contienen suficientes perlas para 2-5 muestras, utilizando al menos 20 perlas de 1,4 mm de diámetro por muestra.
PRECAUCIÓN: El reactivo TRIzol (en adelante, "tampón de homogeneización") es un producto químico peligroso; el contacto con ácidos o lejía libera gases tóxicos; garantizar una ventilación adecuada; Consulte la hoja de datos de seguridad para obtener más información. Si los usuarios sustituyen el reactivo TRIzol o el reactivo TRI por otro tampón de homogeneización, es necesaria una validación adicional. TRIzol actúa como tampón de homogeneización/lisis de muestras, tampón de inactivación de lyssavirus y tampón de estabilidad de ARN para este protocolo. El uso de un tampón de homogeneización alternativo requerirá la validación de la eficiencia de extracción, la inactivación y la estabilidad en una comparación controlada en paralelo. - Limpie y desinfecte la superficie de trabajo con desinfectante de compuestos de amonio cuaternario (QAC) durante 2 minutos y coloque una almohadilla absorbente revestida de plástico. Coloque solo los reactivos y suministros para la primera muestra en un gabinete de seguridad biológica (BSC) de Clase II con características para expulsar humos peligrosos fuera de la habitación.
NOTA: Consulte las instrucciones del fabricante para conocer los límites de almacenamiento de QAC diluido. Asegúrese de que la almohadilla revestida de plástico no bloquee el flujo de aire de la cabina de seguridad biológica. Si se interrumpe el flujo de aire, no use una almohadilla. - Recoja tejido que represente una sección transversal completa del tronco encefálico y el cerebelo utilizando un bisturí limpio de un solo uso.
NOTA: La manipulación de los tejidos debe realizarse de manera que no aerosolice líquidos ni produzca partículas en el aire. No se requieren campanas extractoras ni cabinas de bioseguridad, pero se recomiendan cabinas de bioseguridad ventiladas, ya que brindan protección adicional contra olores, humos, ectoparásitos y fragmentos óseos.
PRECAUCIÓN: El uso de un bisturí con material potencialmente infectado con lyssavirus es peligroso, y los usuarios deben tomar las precauciones de seguridad adecuadas. Se recomienda el uso de pinzas de un solo uso.- En el caso de los animales pequeños (como los murciélagos), se puede recolectar todo el tronco encefálico y el cerebelo.
- En el caso de los animales más grandes, recoja una sección transversal completa del tronco encefálico y el tejido de cada uno de los tres lóbulos del cerebelo.
- OPCIONAL: Si realiza la prueba de anticuerpos fluorescentes directos (DFA), recoja impresiones cerebrales en este punto. Utilice el tejido restante después de recolectar impresiones cerebrales para DFA para la extracción de ARN y las pruebas mediante el ensayo LN34.
NOTA: Si se agrega TRIzol a las muestras, las muestras ya no se pueden usar para métodos de detección basados en antígenos o aislamiento de virus.
- Preparar muestras para la homogeneización y la extracción de ARN.
NOTA: La eficiencia de la extracción de ARN y la inactivación del virus puede verse afectada por el uso excesivo de tejido. La cantidad de tejido no debe exceder aproximadamente 1/10 del volumen del tampón de homogeneización utilizado. Si se utiliza más tejido, aumente la cantidad de tampón de homogeneización en consecuencia para garantizar una extracción de ARN eficiente y exitosa.- En el caso de animales pequeños, coloque todos los tejidos necesarios en un tubo que contenga tampón de homogeneización y perlas para la extracción. No exceda los 100 mg de muestra en 1 mL de tampón de homogeneización; Para muestras más grandes, aumente el volumen del tampón de homogeneización o utilice varios tubos para reflejar una muestra en una proporción de 1:10: tampón.
- En el caso de animales más grandes, pique y homogeneice bien el tejido y retire una porción representativa a un tubo precargado con tampón de homogeneización y perlas. No exceda los 100 mg de muestra en 1 mL de tampón de homogeneización; Para muestras más grandes, aumente el volumen del tampón de homogeneización o utilice varios tubos para reflejar una muestra en una proporción de 1:10: tampón.
- Opción 1 (batidor de cuentas): Homogeneice el tejido con un batidor de cuentas, 1 ml de tampón y cuentas de cerámica. Es posible que sea necesario utilizar varios tubos de 2 ml o tubos más grandes.
- Opción 1 (batidor de cuentas): Limpie y desinfecte la estación de trabajo, el equipo y los tubos de muestra exteriores con desinfectante QAC (1:256). Déjalo reposar durante 2 min.
- Opción 1 (Batidor de cordones): Dentro del BSC, cargue un rotor de centrífuga con muestras homogeneizadas. Centrifugar todas las muestras a 10.000-16.000 × g durante 2 minutos en una microcentrífuga de mesa. Descargue el rotor de la centrífuga dentro del BSC.
- Opción 1 (batidor de cuentas): Déjalo reposar durante 2 min.
- Opción 1 (batidor de cordones): Transfiera 120 μL de homogeneizado a un tubo prellenado con 1 mL de tampón de homogeneización.
PRECAUCIÓN: La homogeneización puede producir aerosoles y debe realizarse en un BSC. - Opción 2 (bisturí): Picar finamente los tejidos necesarios con un bisturí de un solo uso, frotar con un hisopo y transferir el hisopo a un tubo precargado con tampón de homogeneización y perlas. No exceda los 100 mg de muestra en 1 mL de tampón de homogeneización; Para muestras más grandes, aumente el volumen del tampón de homogeneización o utilice varios tubos para reflejar una relación muestra:tampón de 1:10.
PRECAUCIÓN: El uso de un bisturí con material potencialmente infectado con lyssavirus es peligroso, y los usuarios deben tomar las precauciones de seguridad adecuadas.
- Recoja los pañuelos restantes en el recipiente original o en un recipiente nuevo y vacío etiquetado con una etiqueta de acceso. Guarde este tejido en caso de que se requiera una nueva prueba o una caracterización adicional.
- Limpie y desinfecte la estación de trabajo, el equipo y el exterior de los tubos de muestra con el desinfectante QAC 1:256. Déjalo reposar durante 2 min.
- Repita los pasos 1.2 a 1.5 para todas las muestras restantes.
- Homogeneice las muestras con un mini batidor de cuentas durante al menos 60 s. Inspeccione visualmente los tubos. Repita el batidor de cuentas durante 60 segundos adicionales si quedan trozos grandes de pañuelo. Este paso es opcional si el tejido está completamente homogeneizado en el paso 1.4.2.1 anterior.
NOTA: Es importante asegurarse de que el tejido esté completamente homogeneizado. La homogeneización incompleta disminuirá el rendimiento de ARN. - Deje reposar durante al menos 5 minutos a temperatura ambiente (RT).
- Limpie y desinfecte la estación de trabajo, el equipo y el exterior de los tubos de muestra con el desinfectante QAC (1:256).
NOTA: La muestra se considera no infecciosa en este momento y puede extraerse del laboratorio de rabia. - Procese inmediatamente las muestras en tampón de homogeneización para la extracción de ARN, almacene a RT (20 °C a 25 °C) o refrigerado (4 °C a 8 °C) durante varios días, o almacene a -16 °C o más frío para almacenamiento a largo plazo.
2. Protocolo para la extracción de ARN mediante el kit RNA MiniPrep
- Configure el espacio de trabajo en el BSC.
- Limpie la superficie de trabajo de BSC con etanol al 70% antes de comenzar a trabajar para eliminar el polvo u otros contaminantes ambientales. Realice una descontaminación adicional de superficies con el desinfectante QAC (1:256), RNase AWAY o RNaseZap (según las recomendaciones del fabricante).
- Coloque una almohadilla de trabajo absorbente revestida de plástico y coloque los reactivos, los suministros y la muestra en el BSC.
NOTA: Asegúrese de que la almohadilla revestida de plástico no bloquee el flujo de aire del BSC. Si se interrumpe el flujo de aire, no use una almohadilla. - Coloque todos los tubos de recolección en una rejilla limpia para tubos de microcentrífuga. Prellene un tubo de microcentrífuga de 1,5 ml con 300 μl de etanol al 100% para cada muestra de cerebro que no sea de murciélago. Para muestras con poco tejido (muestra de cerebro de murciélago, muestra que no es de cerebro o muestra deteriorada), rellene previamente un tubo de microcentrífuga de 1,5 ml con 600 μl de etanol al 100% para cada uno.
- Preparación de la muestra
- Recoja todas las muestras preparadas en la sección 1 en un estante de tubos en el BSC. Descongele las muestras congeladas justo antes de la prueba.
- Descongelar un control de extracción.
NOTA: Se recomienda elegir una muestra sin ARN de lyssavirus; la muestra debe analizarse previamente con un rango de valores de Ct esperado para el ensayo de beta-actina. Por ejemplo, células de cultivo de tejidos prealícuotas o un caso de rabia negativo previamente analizado (humano o animal).
- Centrifugar todas las muestras a 10.000-16.000 × g durante 2 m en una microcentrífuga de mesa.
- Transfiera el sobrenadante a un nuevo tubo de microcentrífuga estéril que contenga etanol al 100%. Asegúrese de que el sobrenadante sea transparente, sin lípidos evidentes ni tejido sólido. Evite la recolección de lípidos y tejidos sólidos.
- Para tejido cerebral no de murciélago: transferir 300 μL de sobrenadante.
- Para muestras con poco tejido (muestra de cerebro de murciélago, muestra sin cerebro o muestra deteriorada), transfiera 600 μL de sobrenadante.
- Almacene el homogeneizado restante en un tubo de microcentrífuga con tapa de rosca a ≤-16 °C.
- Pipetea hacia arriba y hacia abajo 10 veces para mezclar.
- Para cada muestra, transfiera 600 μL de la mezcla de etanol y sobrenadante a una columna de centrifugación en un tubo de recolección.
- Centrifugar hasta que el líquido haya pasado a través de la columna (1 min a 10.000-16.000 × g). Deseche el flujo continuo.
- Repita si hay más de 600 μL de mezcla de tampón de etanol y homogeneización para una muestra.
- Transfiera cada columna a un nuevo tubo de recolección.
- Añadir 400 μL de tampón de prelavado de ARN a cada columna y centrifugar a 10.000-16.000 × g durante 30 s.
- Deseche el flujo y devuelva cada columna al mismo tubo de recolección.
- Repita los pasos 2.10-2.11.
- Añadir 700 μL de tampón de lavado de ARN a cada columna y centrifugar a 10.000-16.000 × g durante 2 min. Asegúrese de que el tampón de lavado haya pasado por cada columna por completo.
- Transfiera cada columna con cuidado a un tubo libre de RNasa.
- Deseche el flujo continuo y el tubo de recolección de 2.13.
- Añadir 50 μL de agua sin DNasa/RNasa directamente a la matriz de la columna para eluir el ARN.
NOTA: No toque la matriz de la columna con la punta de la pipeta. - Incubar durante 30 s a RT, luego centrifugar a 10.000-16.000 × g durante 1 min.
- Transfiera cuidadosamente el ARN a un nuevo tubo de microcentrífuga marcado con acceso de parte superior plana con tapa de rosca. Mueva el ARN extraído a hielo para realizar pruebas inmediatas. Almacene a largo plazo a -70 °C o menos.
NOTA: El almacenamiento a temperaturas más cálidas o la congelación y descongelación repetidas pueden provocar la degradación del ARN y afectar los resultados del diagnóstico.
3. Protocolo para el ensayo RT-PCR en tiempo real del pan-lyssavirus LN34
- Prepare los reactivos.
- Control positivo artificial
- Si el CDC8 produce un ARN de control positivo artificial, siga las instrucciones del empaque para el almacenamiento, la reconstitución y la alícuota. Omita este paso si ya tiene a mano alícuotas de ARN de control positivo de un solo uso.
NOTA: El ARN de control positivo a concentraciones de trabajo debe manipularse en el área de adición de plantilla y no en la misma área que la preparación de la mezcla maestra. El ARN de control positivo debe producir un valor de umbral de ciclo (Ct) dentro del rango esperado determinado para un lote determinado. Entre corridas, el valor de LN34 Ct para el ARN de control positivo no debe diferir en más de ±1,5 valores de Ct. - Descongele una alícuota de un solo uso del almacenamiento de ≤ -70 °C justo antes de usarla en hielo o bloque de hielo. No congele ni descongele y deseche las alícuotas guardadas durante un tiempo prolongado a temperaturas refrigeradas.
NOTA: Se debe ejecutar un control positivo por triplicado en el ensayo LN34; El control positivo artificial8 no se amplificará en el ensayo de beta-actina.
- Si el CDC8 produce un ARN de control positivo artificial, siga las instrucciones del empaque para el almacenamiento, la reconstitución y la alícuota. Omita este paso si ya tiene a mano alícuotas de ARN de control positivo de un solo uso.
- Control de extracción y muestras: Coloque las muestras recién extraídas en hielo (o bloque de hielo) o descongele las muestras de almacenamiento a ≤ -70 °C en hielo (o bloque de hielo) inmediatamente antes de su uso.
NOTA: El ARN debe descongelarse y procesarse en un área designada para la adición de muestras o plantillas que esté separada de las áreas utilizadas para la preparación de mezclas maestras o la manipulación de productos de PCR o grandes cantidades de material viral (por ejemplo, generación de control positivo, propagación viral)
- Control positivo artificial
- Prepare los reactivos de mezcla maestra en el área de preparación de mezcla maestra.
- Preparación de Mastermix del ensayo RT-PCR LN34 singleplex
NOTA: Los usuarios pueden probar muestras en formato singleplex (paso 3.2.1) o multiplex (paso 3.2.2). No es necesario realizar las versiones 3.2.1 y 3.3.2. La preparación de la mezcla maestra, la alícuota de cebadores y sondas, y los reactivos de control sin plantillas deben descongelarse y manipularse en un área limpia separada del procesamiento de muestras, la necropsia, la PCR y otras áreas donde se manipulan materiales virales. Esto se puede lograr a través de salas separadas o un sistema de gabinete con flujo de muestra unilateral.- Genere mezclas de cebador y sonda a concentraciones de trabajo como se indica en la Tabla 1 y la Tabla 2. Omita este paso si las alícuotas de dilución de trabajo de los cebadores y las sondas ya están a mano.
- Cebadores y sondas alícuotas en existencias de almacenamiento de 1,5 ml y existencias de trabajo de 50 μl y almacene a ≤ -16 °C en la oscuridad. Omita este paso si las alícuotas de dilución de trabajo de los cebadores y las sondas ya están a mano.
NOTA: Se recomienda realizar el paso 3.2.1.2. - Descongele el tampón RT-PCR de un solo paso, sin control de plantilla, cebadores y sondas de almacenamiento de ≤ -16 °C en hielo o bloque de hielo en el área de preparación de la mezcla maestra.
NOTA: Utilice los reactivos hasta la fecha de caducidad o el error de rendimiento, lo que ocurra primero. - Agite brevemente y centrifugue todos los tampones, cebadores y sondas antes de usar.
- Guarde la enzima RT-PCR de un solo paso en hielo o en un bloque de hielo hasta su uso.
- Muestras de ARN
NOTA: Utilice ARN recién extraído siempre que sea posible, ya que la congelación y descongelación puede afectar el rendimiento - Guarde las muestras de ARN en hielo o bloque de hielo hasta su uso.
- Descongele cualquier muestra de ARN congelada en hielo o en un bloque de hielo.
- Etiquete un tubo de microcentrífuga por ensayo (LN34 y βA).
- Determine el número de reacciones (N) que se van a configurar por ensayo.
- Calcule el número de reacciones para el ensayo LN34 multiplicando el número de muestras por 3 y añadiendo 6 para los pocillos de reacción de control más un 10 % de reacciones adicionales para tener en cuenta el volumen perdido durante el pipeteo. (p. ej. para 10 muestras: (10 x 3) + 6 = 36 reacciones; exceso de reacciones: (36 x 0,1) + 36 = 3,6 + 36 = 39,6 reacciones totales, o 40 reacciones redondeadas hacia arriba)
NOTA: Para las pruebas clínicas, se sugiere analizar todas las muestras por triplicado para LN34. A efectos de vigilancia, cada muestra podrá analizarse por duplicado. Se recomienda el uso de triplicado durante la incorporación inicial del ensayo para garantizar una baja variabilidad entre las repeticiones y una buena técnica. - Calcule el número de reacciones para el ensayo βA sumando el número de muestras más 4 pocillos de reacción de control más un 10% de reacciones adicionales para tener en cuenta el volumen perdido durante el pipeteo.
- Determine el volumen de cada reactivo para las mezclas maestras LN34 y βA utilizando la Tabla 2.
- Designe los pocillos para cada muestra que se analizará por triplicado en el ensayo LN34 y singlicar para el ensayo βA utilizando un mapa de placa de 96 pocillos.
- Dispense 23 μL de mezcla maestra de ensayo LN34 en cada pozo de LN34 asignado después de un breve vórtice y centrifugado durante 30 s utilizando una microcentrífuga de mesa para recoger líquido en el fondo del tubo. Evite introducir burbujas.
- Dispense reactivos de 23 μL de mezcla maestra de ensayo βA en cada pozo asignado marcado con βA después de un breve vórtice y centrifugado durante 30 s en una microcentrífuga de mesa para recoger líquido en el fondo.
- Configure las reacciones de control sin plantilla (NTC) pipeteando 2 μL de agua de grado PCR en cada pozo NTC.
- Cubra los pocillos y transfiera la placa al área de adición de la plantilla.
- Realice un vórtice breve y centrifugue los tubos que contienen las muestras de ARN.
- Pipetee 2 μL de ARN extraído de la primera muestra en cada pocillo marcado para esa muestra. Evite introducir burbujas.
- Asegúrese de que el ARN se haya introducido en la pipeta mediante visualización.
- Pipetea en el costado del pocillo para asegurarte de que la muestra se agregue al pocillo correcto.
- Evite agitar las puntas de pipeta que contienen ARN sobre pocillos abiertos tanto como sea posible.
- Repita el paso 3.2.10 para las muestras restantes y el ARN de control positivo.
- Coloque la cubierta adhesiva óptica sobre los pocillos después de agregar todas las muestras y controles. Tenga cuidado de cubrir todos los pozos y sellar completamente.
- Centrifugar a 500 × g durante 1 min a RT en una centrífuga de mesa o use una centrifugadora de placas tipo centrifugadora de ensaladas.
- Coloque la placa sellada en un instrumento de PCR en tiempo real calibrado para tintes reporteros FAM y VIC/HEX y ajústela a los parámetros de ciclo que se muestran en la Tabla 3.
- Preparación de mezclas maestras para el ensayo LN34 Multiplexed (LN34M).
- Etiquete un tubo de microcentrífuga LN34M de acuerdo con la Tabla 2.
- Determine el número de reacciones (N) que se van a configurar por ensayo.
- Calcule el número de reacciones para el ensayo LN34M multiplicando el número de muestras por 3 y agregue 6 para los pocillos de reacción de control más un 10 % de reacciones adicionales para tener en cuenta el volumen perdido durante el pipeteo. (por ejemplo, para 10 muestras: (10 x 3) + 6 = 36 reacciones; exceso de reacciones: (36 x 0,1) + 36 = 3,6 + 36 = 39,6 reacciones totales, o 40 reacciones redondeadas hacia arriba)
- Elija el formato de 25 μL o 12,5 μL. Determine el volumen de cada reactivo para la mezcla maestra LN34M utilizando la Tabla 2.
- Designe los pocillos para cada muestra que se analizará por triplicado en el ensayo LN34M utilizando un mapa de placa de 96 pocillos.
- Dispense reactivos para el ensayo LN34M en los pocillos. Agite brevemente los tubos en vórtice y gire hacia abajo para recoger líquido en el fondo antes de dispensar 23 μL (para una reacción de 25 μL) o 10,5 μL (para una reacción de 12,5 μL) de mezcla maestra en cada pocillo asignado. Evite introducir burbujas.
- Configure las reacciones NTC pipeteando 2 μL de agua de grado PCR en cada pocillo NTC.
- Cubra los pocillos y transfiera la placa al área de adición de la plantilla.
- Brevemente vórtice y gire los tubos que contienen las muestras de ARN para recolectar líquido en el fondo.
- Pipetee 2 μL de ARN extraído de la primera muestra en cada pocillo marcado para esa muestra. Evite introducir burbujas.
- Asegúrese de que el ARN se introduzca en la visualización de la pipeta.
- Pipetea en el costado del pocillo para asegurarte de que la muestra se agregue al pocillo correcto.
- Evite agitar las puntas de pipeta que contienen ARN sobre pocillos abiertos tanto como sea posible.
- Repita el paso 3.3.8 para las muestras restantes y el ARN de control positivo.
- Después de agregar la última muestra/control, coloque la cubierta adhesiva óptica sobre los pocillos, asegurándose de que todos los pocillos estén cubiertos y sellados por completo.
- Centrifugar a 500 × g durante 1 min en RT en una centrífuga de mesa o use una centrifugadora de placas tipo centrifugadora de ensaladas.
- Coloque la placa sellada en un instrumento de PCR en tiempo real calibrado para tintes reporteros FAM y VIC/HEX y ajústela a los parámetros de ciclo, como se muestra en la Tabla 3. Establezca el tinte de referencia pasivo en ROX y ejecute en modo estándar (no se ejecute en modo rápido)
NOTA: Esta configuración es específica de los instrumentos mencionados en este protocolo y requiere el uso de un reactivo RT-PCR de un solo paso que contenga ROX como colorante pasivo. Los instrumentos alternativos requieren diferentes enfoques para determinar los ajustes óptimos de ejecución. Garantice el mantenimiento normal del instrumento según el fabricante para obtener el mejor rendimiento.
- Preparación de Mastermix del ensayo RT-PCR LN34 singleplex
4. Interpretación de los resultados
- Establezca una línea de base automática y cálculos de umbral manuales utilizando un valor de 0,2 para LN34/FAM y 0,05 para βA/HEX/VIC.
NOTA: Esta configuración es específica de los instrumentos mencionados en este protocolo y requiere el uso de un reactivo RT-PCR de un solo paso que contenga ROX como colorante pasivo. Los instrumentos alternativos requieren diferentes enfoques para calcular los valores de referencia y umbral. - Determine el resultado del diagnóstico utilizando la guía de la Tabla 4 si todos los controles funcionaron como se esperaba (Tabla 5).
- Confirme todos los valores de Ct o Cq mediante la visualización de gráficos de amplificación.
- Investigue cualquier resultado inusual como se recomienda.
5. Retención y almacenamiento de muestras
- Almacene todas las muestras congeladas a -16 °C o menos hasta que se completen las pruebas y se informen los resultados. Conserve los tejidos originales para confirmar los resultados o identificar al animal huésped de la especie en caso de resultados inusuales en las pruebas.
- Utilice identificadores de muestra únicos; Etiquete todos los tubos, informes y documentos con identificadores de muestra únicos completos.
- Conserve las muestras intermedias (a corto plazo) en caso de que se requiera repetir la prueba.
- Conserve muestras positivas representativas según sea necesario para su uso como controles, tipificación epidemiológica y otros fines.
- Almacene el ARN a ≤-70 °C para su almacenamiento a largo plazo.
Resultados
En la Figura 2 se muestran imágenes representativas de un ensayo LN34 ejecutado con éxito en un instrumento de PCR en tiempo real ABI ViiA7. La visualización de los resultados trazados en una escala logarítmica permite una fácil visualización del valor Ct, el punto en el que la curva cruza la línea de umbral (Figura 2A, C). Cuando se traza en una escala lineal, la amplificación exitosa aparecerá como una curva sigmoidal (o en forma de "S") (Figura 2B, D), mientras que los resultados negativos deben aparecer como una línea recta y plana. Se recomienda ver los resultados en las vistas de escala lineal y logarítmica para identificar posibles anomalías o errores. Los resultados positivos y negativos típicos en la vista de gráfico multicomponente se pueden ver en la Figura 2E, F, respectivamente, donde se puede observar el nivel de fluorescencia del colorante que marca la sonda (FAM para LN34, VIC/HEX para βA) en relación con el colorante pasivo en el tampón de reacción (ROX).
En la Figura 3 se muestran ejemplos de resultados anormales. Las comparaciones entre los gráficos de ejecuciones exitosas (Figura 2) y los gráficos anormales (Figura 3) se pueden utilizar para aislar ejecuciones atípicas y problemas de instrumentos. La Figura 3A muestra una señal que cruza el umbral, produciendo un valor de Ct para LN34, pero la curva de amplificación es muy atípica, aumentando linealmente. El gráfico multicomponente (Figura 3B) también muestra una línea ondulada que no es típica de una muestra positiva. Este ejemplo resalta la importancia de ver los gráficos de amplificación y no simplemente copiar los valores de Ct. Asegúrese siempre de que las curvas de amplificación se vean normales para todas las muestras. También se recomienda ver el gráfico multicomponente para asegurarse de que no haya irregularidades. En ocasiones, las señales de referencia desordenadas pueden generar valores de Ct en casos en los que no se ha producido ninguna amplificación. Si las señales de amplificación parecen lineales, se sugiere ajustar la línea de base para ver si la curva desaparece. En el caso de cualquier señal inusual, se debe repetir toda la ejecución. Se recomienda limpiar y ejecutar una placa de fondo en su instrumento de PCR en tiempo real si los problemas persisten. Si están disponibles, los productos de PCR se pueden ejecutar en un gel de agarosa y/o secuenciar para solucionar cualquier resultado inusual. No se recomienda utilizar los resultados de la electroforesis en gel o la secuenciación para determinar los resultados del diagnóstico.
Estudios previos han demostrado una baja variabilidad entre las réplicas, la ejecución del ensayo, el operador y el laboratorio para el ensayo LN347. Si se observa una alta variabilidad (diferencia de >±1,5 Ct) entre las réplicas de la misma muestra, se debe volver a analizar ese ARN. La alta variabilidad puede deberse a problemas con las pipetas, las prácticas de laboratorio, el pipeteo incorrecto o las máquinas de PCR en tiempo real. La observación repetida de una alta variabilidad entre varias muestras o entre series de ensayos puede indicar problemas sistémicos. Las muestras con ARN bajo, que se acercan al umbral de ensayo para una muestra positiva (Ct 35), pueden exhibir una mayor variabilidad en los valores de Ct entre las réplicas. Es posible que sea necesario consultar con los CDC y solucionar problemas para abordar la causa de la variabilidad persistente, los resultados inconsistentes o la falla del ensayo.
La alta sensibilidad de los ensayos basados en PCR los hace inherentemente susceptibles a la contaminación. El cumplimiento estricto de las buenas prácticas de laboratorio es la mejor manera de mitigar la contaminación cruzada. Es importante saber cómo identificar la contaminación potencial. Se debe sospechar de contaminación del reactivo si no hay un control de plantilla y se sospecha que los pocillos de muestra negativos en una ejecución de ensayo producen valores de Ct similares. Repita las pruebas con nuevas alícuotas de reactivos de PCR (tampón, agua, cebadores y enzima) y el mismo ARN. Si todas las muestras y el control de extracción producen valores similares de CT pero NTC es negativo, se debe investigar la contaminación de los reactivos de extracción y se debe repetir la extracción con nuevos reactivos. Es una buena práctica hacer pequeñas alícuotas de reactivos para reducir el riesgo de contaminación y evitar la posibilidad de descartar grandes volúmenes de reactivos costosos. La contaminación cruzada de la muestra es más difícil de identificar. Si se sospecha de contaminación de la muestra, repita la recolección de la muestra comenzando desde los tejidos originales. En algunos casos, la secuenciación del ARN viral puede confirmar la contaminación, especialmente cuando el ARN contaminante es muy diferente de la variante viral esperada (como un virus de control utilizado en el laboratorio). La secuenciación de dos muestras procesadas al mismo tiempo puede determinar si las secuencias virales son idénticas, pero puede ser poco informativa si se espera que las secuencias sean muy similares (por ejemplo, la misma variante recolectada en el mismo condado). Si se sospecha de contaminación de la muestra con el ARN de control positivo, se pueden ejecutar los amplicones del ensayo LN34 en un gel de agarosa para diferenciar el ARN del lisavirus (165 pb) del ARN de control positivo (99 pb). La secuencia del molde utilizado para generar el ARN de control positivo proporcionado por CDC8.
Para otros patógenos, se pueden usar laboratoristas para establecer el umbral manualmente para eliminar el "ruido", como la amplificación débil que se muestra en cian en la Figura 4. Esta práctica NO se recomienda para el diagnóstico de la rabia porque puede dar lugar a resultados falsos negativos con consecuencias nefastas, ya que la rabia es casi 100% mortal. NO cambie manualmente el umbral para producir resultados negativos para muestras de amplificación débil o tardía. Estas muestras deben ser extraídas y/o analizadas de nuevo para descartar la rabia.
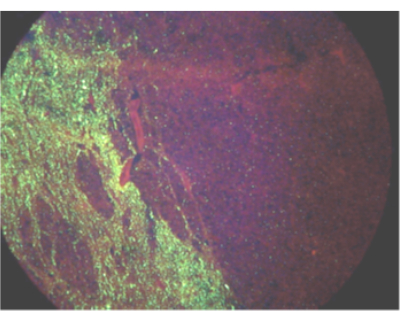
Figura 1: Campo de visión que muestra la propagación unilateral del antígeno del virus de la rabia en un burro infectado mediante una prueba de anticuerpos fluorescentes directos. Haga clic aquí para ver una versión más grande de esta figura.
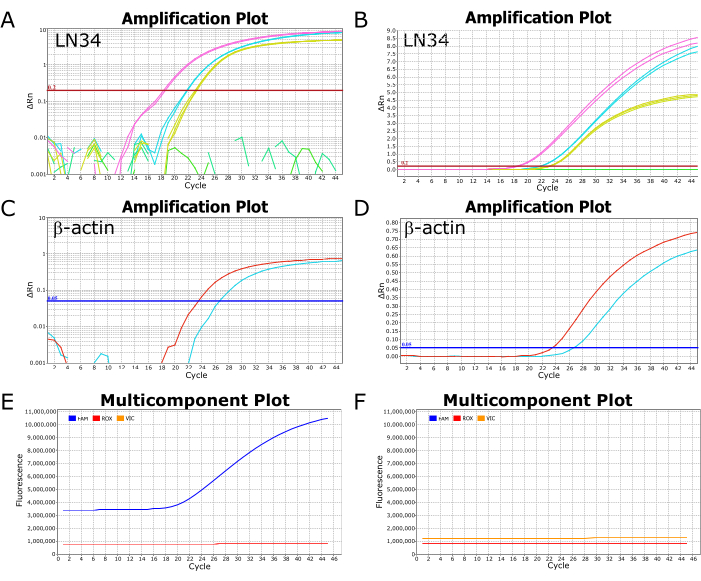
Figura 2: Gráficos de amplificación y multicomponente de una ejecución exitosa del ensayo LN34. (A-D) Los datos de los resultados se trazan en una escala logarítmica (A,C) y una escala lineal (B,D) para el ensayo LN34 y βA. Los paneles A y B representan los resultados de LN34 de dos muestras (en rosa y cian) en comparación con el control positivo (en amarillo). En el panel B, hay una línea verde plana que representa una muestra negativa adicional en la ejecución. En A, la(s) línea(s) verde(s) no muestra(n) ninguna amplificación y se representan como segmentos rotos. El umbral para el ensayo LN34 se estableció manualmente en 0,2 y se muestra mediante una línea horizontal roja. (C,D) Resultados del ensayo βA para dos muestras (rojo y cian). El umbral para el ensayo βA se estableció manualmente en 0,05. (E,F) Los gráficos multicomponente representan la fluorescencia (RFU) en cada ciclo para FAM (LN34), VIC (βA) y ROX (colorante pasivo presente en el tampón AgPath-ID). Los niveles de ROX deben permanecer estables a lo largo de todos los ciclos. En el panel E se muestra una muestra positiva típica; La fluorescencia FAM aumenta como una curva sigmoidal a partir del ciclo 18 para esta muestra. Una muestra negativa típica se muestra en el panel F, donde el nivel de FAM permanece paralelo al nivel de ROX a lo largo de todos los ciclos. Los datos provienen de un instrumento de PCR en tiempo real ABI ViiA7. Haga clic aquí para ver una versión más grande de esta figura.

Figura 3: Imágenes representativas de la señal rara y atípica observada en las ejecuciones del ensayo LN34 en un instrumento de PCR en tiempo real ViiA7. (De la A a la F) Parcelas de amplificación (A,C,E) y multicomponente (B,D,F) producidas debido a la contaminación del pozo. El aumento lineal (A,C) y las fluctuaciones onduladas (B,D) en la fluorescencia de FAM no representan una verdadera amplificación basada en la forma de las curvas y la magnitud del cambio de fluorescencia. Es probable que los paneles A a D representen muestras negativas aunque se haya producido un valor de Ct para la réplica que se muestra en los paneles A y B. Los paneles E y F muestran una extraña señal ondulada que se ve más fácilmente en el gráfico multicomponente. Este tipo de señal debe investigarse y puede indicar problemas con el instrumento, aunque todos los controles se hayan realizado como se esperaba en esta ejecución. Haga clic aquí para ver una versión más grande de esta figura.
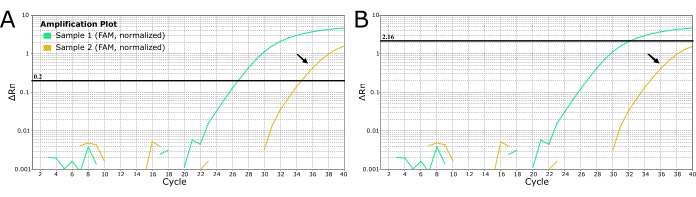
Figura 4: Curvas de RT-PCR en tiempo real de LN34 de 2 muestras sospechosas de rabia que muestran dos métodos para establecer los valores umbral. (A) El umbral de LN34 se estableció en 0,2 (recomendado para todas las corridas). (B) Determinación manual de un umbral diferente para cada corrida para enmascarar la señal que se determine como "ruido" (señal de amplificación tardía). El método utilizado en el panel B NO se recomienda para la rabia debido a las graves consecuencias de omitir un resultado positivo verdadero. La amplificación tardía podría indicar una muestra positiva débil, inhibición de la PCR o una extracción fallida en un caso positivo. También podría indicar contaminación cruzada. La muestra áurea (indicada con flechas negras) produce un valor de Ct en el punto de corte del ensayo y no debe considerarse negativa. Las muestras con amplificación tardía deben volver a extraerse y volver a analizarse. Haga clic aquí para ver una versión más grande de esta figura.
Tabla 1: Secuencias y concentraciones de cebadores y sondas utilizadas en los ensayos de RT-PCR en tiempo real LN34lys (LN34 singleplex LN34), LN34M (LN34 y βA multiplexados). Las sondas LN34 están marcadas con el colorante fluorescente FAM en el extremo 5 y el extintor de agujeros negros (BHQ1) en el extremo 3. La sonda βA está marcada con el tinte fluorescente HEX en el extremo 5 y el extintor de agujeros negros (BHQ1) en el extremo 3. Las bases bloqueadas modificadas con nucleótidos se indican mediante un signo más que precede a la base en la secuencia. Haga clic aquí para descargar esta tabla.
Tabla 2: Configuración del ensayo para los ensayos LN34lys, Actin3 y LN34M. Los nombres, secuencias y concentraciones de cebadores y sondas se pueden encontrar en la Tabla 1. LN34_F1 corresponde a ACGCTTAACAACCAGATCAAAGAA7. Haga clic aquí para descargar esta tabla.
Tabla 3: Parámetros de ciclo para instrumentos ABI. IMPORTANTE: Asegúrese de ejecutar en modo ESTÁNDAR , no en modo RÁPIDO . ROX debe seleccionarse como el tinte de referencia pasivo. Haga clic aquí para descargar esta tabla.
Tabla 4: Algoritmo para la interpretación de los resultados de RT-PCR en tiempo real de LN34 para formatos singleplex (arriba, tabla azul) y multiplex (abajo, tabla roja). Un resultado positivo de LN34 debe considerarse positivo, incluso si el resultado de βA es negativo o no concluyente. Si no se detecta el amplicón LN34, el βA Ct debe estar ≤ el valor de corte de Ct indicado para que se considere negativo. Los valores de βA Ct indican la calidad de la muestra que se está analizando e identifican una posible inhibición. La baja concentración en el espécimen clínico original puede afectar las curvas de crecimiento de βA, lo que no conduce a una amplificación discernible. Otros factores que contribuyen a la imposibilidad de detectar β-actina son la mala extracción de ARN debido a la pérdida de ARN o al arrastre de inhibidores de la PCR, la configuración y la técnica incorrectas del ensayo, el tipo o la calidad de la muestra insatisfactorios y el mal funcionamiento de los reactivos o el equipo. Haga clic aquí para descargar esta tabla.
Tabla 5: Acciones e interpretaciones de los resultados comunes para los controles del ensayo LN34. Los tres controles (ARN de control positivo de la rabia, control negativo de extracción de la rabia y sin control de plantilla) deben producir los resultados esperados para que una tirada pase. La falla en el control positivo o la ausencia de control de plantilla puede indicar un error de pipeteo, reactivo o falla del equipo. Se debe repetir toda la ejecución, incluidas todas las muestras de ARN analizadas. Un fallo en el control de extracción puede indicar un problema durante la extracción, como un fallo del reactivo, un pipeteo incorrecto o una contaminación cruzada. La extracción de todas las muestras debe repetirse. El fracaso de los controles debería ser raro para el personal de laboratorio experimentado. Haga clic aquí para descargar esta tabla.
Discusión
Una ejecución exitosa del ensayo LN34 requiere un control positivo, un control de extracción, y ninguna reacción de control de molde se comporta como se espera en cada ejecución del ensayo o la ejecución debe invalidarse y repetirse. Las tres reacciones replicadas de control positivo LN34 deben cruzar el umbral dentro del rango especificado, o se debe repetir la ejecución. El ARN de control positivo descrito en publicaciones anteriores 7,8 no se amplificará en el ensayo βA. Las reacciones de control sin molde no deben exhibir curvas de amplificación que crucen la línea de umbral para el ensayo LN34 o βA. El control de extracción no debe exhibir amplificación para LN34. Si se observa una amplificación inesperada en el NTC o en el control de extracción, puede indicar contaminación e invalidar la ejecución y repetición de la prueba para todas las muestras (ver Tabla 5). Los usuarios pueden considerar agregar controles adicionales, incluido un control sin proceso o un control sin extracción de muestras, para monitorear la contaminación del huésped βA de los reactivos de extracción.
A medida que la mortalidad por rabia se acerca al 100%, se recomienda que cualquier amplificación débil o anormal se investigue más a fondo, incluso si no produce un valor de Ct. Las reacciones negativas o NTC no deben exhibir ninguna amplificación, y la fluorescencia debe aparecer como una línea plana paralela a la fluorescencia ROX en la vista multicomponente. Las observaciones de curvas, especialmente en múltiples réplicas, pueden indicar contaminación cruzada o un resultado positivo débil. Todas las réplicas de una muestra positiva deben amplificarse para obtener un resultado positivo válido. Si solo un subconjunto de réplicas se amplifica en cualquiera de los ensayos, se debe volver a analizar la muestra. Además, cualquier muestra que produzca resultados muy variables (diferencias en el valor de Ct > ±1,5 entre las réplicas) debe considerarse inválida y la muestra debe volver a analizarse. Si el problema persiste, se debe volver a extraer la muestra.
Se espera que una muestra positiva para la rabia extraída de tejido del tronco encefálico y del cerebelo recolectada y almacenada correctamente tenga un valor de Ct de menos de 35 ciclos para el ensayo LN34. Todas las muestras no concluyentes deben volver a analizarse mediante RT-PCR en tiempo real LN34. Si la muestra no es concluyente después de repetir la prueba y todos los controles se realizaron como se esperaba, se recomienda volver a extraer el ARN. Las muestras con ARN viral bajo (LN34 Ct > 35) pueden indicar problemas potenciales como contaminación, baja carga viral, inhibición de PCR o extracción fallida. Recoja piezas frescas del cerebro del tejido original, realice la extracción de ARN y vuelva a analizar la muestra. Del mismo modo, los valores de Ct > 33 (singleplex), 37 (LN34M) o ninguna amplificación en el ensayo βA pueden indicar una extracción fallida de ARN. Repita la extracción de dichas muestras y, a continuación, repita las pruebas de LN34 y actina. Si una muestra vuelve a producir un resultado no concluyente después de repetidas pruebas, utilice un método secundario como la prueba DFA (también llamada FAT), DRIT o aislamiento de virus. Si se observan resultados discordantes continuos o resultados no concluyentes, consulte con un laboratorio de referencia de rabia para realizar pruebas confirmatorias.
Si los inhibidores están presentes en una extracción de ARN, los ensayos de PCR pueden producir un resultado falso negativo. Si se sospecha de inhibición o se observa inhibición de las reacciones de control βA (como el valor Ct > 33 o el valor Ct > 37) para una muestra en particular, el ARN extraído debe analizarse en 2 o más diluciones (por ejemplo, 1:10 y 1:100 en agua libre de nucleasas) para diluir cualquier posible inhibidor de PCR. Para muestras difíciles, la entrada de ARN se puede aumentar a 8,5 μL en la reacción RT-PCR sin agregar agua. Esto puede revelar un aumento de la inhibición (un valor de Ct más tardío en comparación con el ARN de entrada de 2 μL) o un nivel bajo de ARN en la muestra original (un valor de Ct más temprano cuando se utilizan 8,5 μL en comparación con el ARN de entrada de 2 μL).
El ensayo LN34 no diferencia entre lyssavirus ni determina variantes del virus de la rabia. El amplicón del ensayo LN34 se puede secuenciar para la tipificación de variantes del virus de la rabia de baja resolución o la identificación de especies de lyssavirus11.
Divulgaciones
Ninguno para revelar
Agradecimientos
Reconocemos los esfuerzos y la colaboración de muchos laboratorios de pruebas diagnósticas de rabia que han contribuido a la implementación, validación y optimización del ensayo LN34 a través de su intercambio de datos abiertos y comentarios. El uso de nombres comerciales y fuentes comerciales es solo para identificación y no implica la aprobación de los Centros para el Control y la Prevención de Enfermedades, el Departamento de Salud y Servicios Humanos de EE. UU. o las instituciones afiliadas a los autores. Las conclusiones, hallazgos y opiniones expresadas por los autores no reflejan necesariamente la posición oficial del Departamento de Salud y Servicios Humanos de los Estados Unidos, los Centros para el Control y la Prevención de Enfermedades o las instituciones afiliadas a los autores.
Materiales
| Name | Company | Catalog Number | Comments |
| 7500 Fast | Applied Biosystems | N/A | Do not substitute without validation |
| 7500 Fast Dx | Applied Biosystems | N/A | Do not substitute without validation |
| ABI ViiA 7 | Applied Biosystems | N/A | Do not substitute without validation |
| AgPath-ID One-Step RT-PCR Kit | ThermoFisher Scientific | AM1005 | Do not substitute without validation |
| Beadbug6 | Benchmark Scientific | D1036 | |
| Direct-zol RNA MiniPrep kit | Zymo Research | R2052 | |
| MagNA Lyser green beads | Roche | 3358941001 | |
| Microcentrifuge | Eppendorf | 5425 R | |
| Optical 96-well Reaction Plates | ThermoFisher Scientific | 4346907 | |
| Optical Adhesive covers | ThermoFisher Scientific | 4311971 | Alternative: caps |
| Polyester fiber-tipped applicator swabs | BD BBL Polyester Fiber Tipped Application Swab | 220690 | |
| QuantStudio 6Flex | Applied Biosystems | 4485691 | Do not substitute without validation |
| Quaternary ammonium disinfectant (1:256) | LYSOL | WBB56939 | Do not substitute without validation |
| RNase AWAY | ThermoFisher Scientific | 7002PK | |
| RNaseZap | ThermoFisher Scientific | AM9780 | |
| Single-use scalpel, a scalpel with a safety mechanism | Integra Miltex | 4-510 | |
| Sterile polyproylene microcentrifuge tubes (1.5 mL), nuclease free | Sarstedt | 72.692.405 | |
| Sterile polyproylene microcentrifuge tubes (2 mL), nuclease free | Sarstedt | 72.694.600 | |
| TRIzol Reagent | ThermoFisher Scientific | 15596026 | Do not substitute without validation |
Referencias
- Meechan, P. J., Potts, J. Biosafety in Microbiological and Biomedical Laboratories. , Centers for Disease Control and Prevention. (2020).
- Terrestrial Manual 2023. , World Organization for Animal Health. At https://www.woah.org/en/what-we-do/standards/codes-and-manuals (2023).
- Laboratory Techniques in Rabies. Volume 1, World Health Organization. At https://iris.who.int/handle/10665/310836 (2018).
- Laboratory Techniques in Rabies. Volume 2, World Health Organization. At https://iris.who.int/bitstream/handle/10665/310837/9789241515306-eng.pdf?ua=1 (2019).
- Genevie, R., et al. Protocol for postmortem diagnosis of rabies in animals by direct fluorescent antibody testing: A minimum standard for rabies diagnosis in the United States. , At https://www.cdc.gov/rabies/pdf/RabiesDFASPv2.pdf (2003).
- World Health Organization. WHO Expert Consultation on Rabies: Third Report. , World Health Organization. Geneva. (2018).
- Gigante, C. M., et al. Multi-site evaluation of the LN34 pan-lyssavirus real-time RT-PCR assay for postmortem rabies diagnostics. PLoS One. 13 (5), e0197074(2018).
- Wadhwa, A., et al. A Pan-Lyssavirus Taqman Real-Time RT-PCR assay for the detection of highly variable rabies virus and other lyssaviruses. PLoS Negl Trop Dis. 11 (1), e0005258(2017).
- Gigante, C. M., Wicker, V., Wilkins, K., Seiders, M., Zhao, H., Patel, P., Orciari, L., Condori, R. E., Dettinger, L., Yager, P., Xia, D., Li, Y., et al. Optimization of pan-lyssavirus LN34 assay for streamlined rabies diagnostics by real-time RT-PCR. Journal Virological Methods. , In press (2024).
- Rao, A. K., et al. Use of a modified preexposure prophylaxis vaccination schedule to prevent human rabies: recommendations of the advisory committee on immunization practices-United States, 2022. Morbidity and Mortality Weekly Report. 71 (18), 619(2022).
- Condori, R. E., et al. Using the LN34 Pan-Lyssavirus Real-Time RT-PCR assay for rabies diagnosis and rapid genetic typing from formalin-fixed human brain tissue. Viruses. 12 (1), 120(2020).
Reimpresiones y Permisos
Solicitar permiso para reutilizar el texto o las figuras de este JoVE artículos
Solicitar permisoExplorar más artículos
This article has been published
Video Coming Soon
ACERCA DE JoVE
Copyright © 2025 MyJoVE Corporation. Todos los derechos reservados