このコンテンツを視聴するには、JoVE 購読が必要です。 サインイン又は無料トライアルを申し込む。
Method Article
RGBradford:スマートフォンカメラによるタンパク質定量(英語)
要約
この論文では、ブラッドフォードアッセイとスマートフォンを分析デバイスとして使用したタンパク質定量のためのプロトコルを提供します。サンプル中のタンパク質レベルは、スマートフォンで撮影したマイクロプレートの写真から抽出したカラーデータを使用して定量化できます。
要約
タンパク質の定量は、ライフサイエンス研究において不可欠な手順です。他のいくつかの方法の中でも、ブラッドフォードアッセイは最も使用されている方法の1つです。ブラッドフォードアッセイは広く普及しているため、その性能を向上させるための元の方法のいくつかの変更を含め、限界と利点が徹底的に報告されています。元の方法の変更点の1つは、分析機器としてスマートフォンのカメラを使用することです。この論文では、ブラッドフォードアッセイの条件に存在する3種類のクマシーブリリアントブルー色素を利用して、マイクロプレートの1枚の写真から抽出したカラーデータを使用して、サンプル中のタンパク質を正確に定量する方法について説明します。マイクロプレートでアッセイを行った後、スマートフォンのカメラで写真を撮影し、その画像から無料のオープンソースの画像解析ソフトウェアアプリケーションを使用してRGBカラーデータを抽出します。次に、タンパク質濃度が不明なサンプルの青と緑の強度の比率(RGBスケール)を使用して、標準曲線に基づいてタンパク質含有量を計算します。RGBカラーデータを用いて計算した値と従来の吸光度データを用いて計算した値との間に有意差は認められなかった。
概要
下流の用途(ELISA、酵素動態、ウェスタンブロッティング、タンパク質精製、質量分析など)にかかわらず、ライフサイエンスラボでの正確な分析にはタンパク質の定量が不可欠です。二次読み出し(タンパク質の質量あたりの分析種の相対レベルを計算する)としての使用に加えて、サンプル中のタンパク質レベルは、それ自体が目的の出力になることもあります。例えば、食物資源1や尿2のタンパク質レベルに関心を持つことができます。サンプル3のタンパク質濃度の測定には、UV吸光度の直接測定4、タンパク質-銅キレート化5,6、タンパク質-色素結合比色アッセイ7、タンパク質-色素結合蛍光アッセイ8など、多くの方法があります。タンパク質定量の関連性は、最も引用された文献9,10のトップ3にタンパク質測定法5,7を説明する2つの論文が存在することによって証明されています。多くの著者が、一次文献以外の文献を引用したり、何も引用しなかったりして、実際の引用を無視しているという事実にもかかわらず、Lowryタンパク質アッセイとブラッドフォードタンパク質アッセイを記述した元の論文は、それぞれ200,000>引用されています10。
ブラッドフォードアッセイの人気は、その手頃な価格、シンプルさ、スピード、感度に起因しています。このアッセイは、酸性条件下でのタンパク質と色素Coomassie Brilliant Blue Gとの相互作用に基づいています。アッセイの条件下(すなわち、低pH)では、色素は3つの形態で存在します:470 nmでλmaxを有する赤色のカチオン型。650 nmにλmaxを有する緑色の中性体。λmaxが590 nm11,12の青色アニオン型です(図1)。カチオン型は、タンパク質の非存在下で優勢である。タンパク質が色素と相互作用すると、青色の陰イオン性形態が安定化し、溶液の色が茶色がかった色から青色に顕著な変化が生じます。通常、青色の色素の濃度の変化は分光光度法で定量化され、その590〜595nmでの吸光度はアッセイ中のタンパク質の量に比例します。
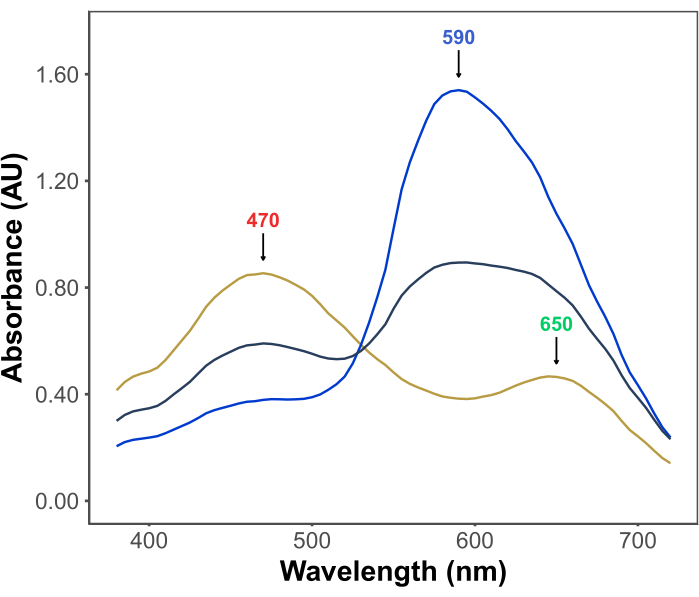
図1:ブラッドフォードアッセイの条件下でのクマシーブリリアントブルーG吸収スペクトル。 3 つの主要なピークには、赤色(470 nm)、緑色(650 nm)、青色(590 nm)の色素の λmax を示す矢印が付いています。スペクトルは、タンパク質の非存在下(黄色の線)および3 μg(灰色の線)および10 μg(青の線)のウシ血清アルブミンの存在下で記録されました。 この図の拡大版をご覧になるには、ここをクリックしてください。
ブラッドフォードアッセイの広範な使用は、いくつかの制限(例えば、異なるタンパク質に対するさまざまな応答11、脂質13と界面活性剤7による干渉)の特定と、その性能を改善するための修飾の開発(例えば、界面活性剤の添加14,15、アルカリ化14,16、吸光度の比率の使用17)の開発につながった).アッセイ自体の変更に加えて、分析シグナルを捕捉するためのスマートフォンやカメラなどの代替デバイスの使用も説明されています18,19,20。実際、スマートフォンを携帯型化学分析装置として活用する方法の開発は、活発な研究分野です。スマートフォンを使用する動機は、これらのデバイスの手頃な価格、携帯性、使いやすさ、および広範な可用性に起因しています。
この論文では、スマートフォンを分析デバイスとして使用するRGBradfordアッセイ20を使用したタンパク質定量のためのプロトコルを提供します。元のRGBradford公報20とは対照的に、ここでは、色抽出プロセスを合理化する手順が導入されている。これは、無料で入手できるソフトウェアアプリケーションを利用して、マイクロプレート画像の各ウェルから色情報を自動的に抽出し、時間と労力を大幅に節約することを含みます。これは、グラフィックエディタソフトウェアアプリケーション20を用いて、各ウェルから色データを1つずつ手動で取得する以前の方法に代わるものである。最終的には、スマートフォンで撮影したマイクロプレートの写真から抽出したカラーデータを使用して、サンプル中のタンパク質レベルを定量化できます。
プロトコル
1. ブラッドフォードタンパク質アッセイ試薬の調製
- 100 mgのクマシーブリリアントブルーGを50 mLの95%(w/v)エタノールに溶解します。クマシーブリリアントブルーGが完全に溶けるまで混ぜます。
注意: エタノールは可燃性であり、目の炎症を引き起こします。炎を避け、ゴーグルを使用してください。 - 前の溶液に、100 mLの85%(w / v)リン酸を注意深く加えます。
注意: リン酸は金属を腐食し、皮膚の腐食、深刻な眼の損傷、および急性経口毒性を引き起こします。手袋とゴーグルを着用してください。 - クマシーブリリアントブルーG、エタノール、リン酸を含む溶液を600 mLの脱イオン水にゆっくりと加えます。
- 溶液を最終容量1,000 mLに希釈します。分析するサンプルの数に応じてスケールアップまたはスケールダウンします。元のメソッド7 に記載されているように、作業中の Bradford 試薬の最終濃度は、0.01% (w/v) の Coomassie Brilliant Blue G、4.7% (w/v) のエタノール、および 8.5% (w/v) のリン酸です。
- ろ紙(Whatman#1紙または同等品)でろ過された不溶性材料を取り除きます。
- この試薬は、室温(RT)で保存し、光から保護すると、数週間安定しています。時間の経過とともに沈殿物が形成される可能性があるため、必要に応じてろ過してください。
注:あるいは、すぐに希釈して使用できるブラッドフォード試薬が市販されています。試薬を調製するための製造元の指示に従い、次のステップに進みます。
2. 蛋白質標準液の調製
- 標準として使用する単離タンパク質のストック溶液(ステップ2.4を参照)を調製します。手頃な価格で一般的に使用されているタンパク質は、ウシ血清アルブミン(BSA)です。他の選択肢は、オボアルブミンとウシガンマグロブリンです。
- 標準物質として用いるタンパク質のモル吸収率がわかっている場合は、分光光度計で原液の濃度を確認してください。
- BSA の場合、一般的に使用される式は BSA (mg/mL) = (A280/6.6) × 10 であり、ここで A280 は、適切なブランクに対して読み取られた光路長 1 cm の 280 nm での吸光度です(つまり、ε2801% = 6.6)7。例えば、0.8 mg/mL BSA の吸光度は 280 nm で 0.528 です。
- 検量線を生成するには、0.025 mg/mL から 1.0 mg/mL の範囲で BSA を数倍希釈します。これらは、ウェルあたり10μLのサンプル量を追加した後、ウェルあたり0.25〜10μgのBSAになります。
注:タンパク質標準溶液は、サンプルの調製に使用した培地と同じ組成(最終濃度)の培地で調製する必要があります。
3. アッセイ
- サンプルを希釈して、タンパク質濃度が標準曲線の 0.025-1.0 mg/mL の範囲内に収まるようにします。範囲内に複数(少なくとも 3 つ)のサンプル希釈がある。
注:サンプルは、リン酸緩衝生理食塩水(PBS)またはブラッドフォード試薬と互換性のあるその他の培地/緩衝液組成物で希釈できます。培地成分の最終濃度は、標準試料とサンプルで異なるべきではありません。 - 各タンパク質標準溶液 10 μL を 96 ウェルマイクロプレートの 3 ウェル(3 ウェル)に添加します。タンパク質ポイントが 0(ゼロ)の場合は、標準溶液の調製に使用したバッファー/培地を 10 μL 添加し、サンプルを希釈します。
- 別のウェルセットに、同じ 96 ウェルマイクロプレートの 3 つのウェル(つまり、3 回に分けて)に希釈した各サンプル 10 μL を加えます。1つのアプローチは、異なる容量のサンプルを添加し、ウェルあたり最大10 μLの培地(例えば、0.5 μL、1 μL、2 μL、4 μL、8 μLのサンプルと、9.5 μL、9 μL、8 μL、6 μL、2 μLの培地)で完成させることです。
- 250 μLのBradfordタンパク質アッセイ試薬をすべてのウェルに加えます。典型的なマイクロプレートのセットアップを 図 2 に示します。この例では、260 μL(ウェルあたりの最終容量)の水を含むブランクウェルのセットを、マイクロプレートリーダーのブランクとして含めました(ステップ4.5)。これは、吸光度データも収集する場合にのみ必要です。
注:アッセイを行うたびに、サンプルの同じマイクロプレートに検量線を調製してください。つまり、サンプル数に応じて別のプレートが必要な場合は、2枚目のプレートに別の検量線を用意します。 - 5〜15分以内に結果(セクション4)を記録します。
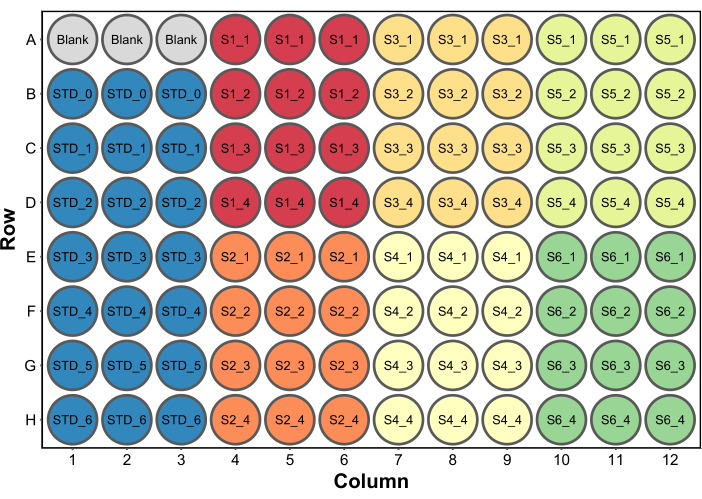
図2:Bradfordタンパク質アッセイの典型的なプレートレイアウト。 ブランクとは、マイクロプレートリーダーでブランクとして使用する260μLの水を含む3つのウェルを指します。STDとは、タンパク質の標準物質を指します。S1-S6 は 6 つの異なるサンプルです。SX_1-SX_4は、サンプルごとに 4 つの異なるサンプル希釈液です。 この図の拡大版をご覧になるには、ここをクリックしてください。
4. 結果の記録
- 明るい部屋で、マイクロプレートをベンチと平行に、均一な白い背景(紙シートなど)を片手で持ちます。プレートに水準器を配置して、正確な位置合わせを確保します。
- 一方、スマートフォンをベンチとマイクロプレートに平行に持ち(一部のカメラアプリケーションはデバイスの傾きを便利に示します)、マイクロプレート全体の写真を1枚または数枚撮影します(図3)。iOSデバイスでは、 グリッド オプションを有効にして、カメラ設定でカメラレベルインジケーターをオンにします。Android デバイスでは、 フレーミング ヒントを有効にして、カメラ設定でカメラ レベル インジケーターをオンにします。
- 特別な照明器具は必要ありませんが、影や反射に注意してください。例えば、スマートフォンでプレートや背景を陰影化したり、マイクロプレートで背景を陰影化したりしないでください。井戸領域の端の小さな反射は問題ではありません。カラーデータは、各ウェルの中央にある非常に小さな領域から抽出できます。
- 背景の均一性、影、反射について画像を簡単に確認します。また、井戸の角度を見てください。各井戸の中心は、直接見えるようにする必要があります(つまり、井戸の壁の後ろではありません)。
- マイクロプレートの吸光度測定値とピクチャーカラーデータの比較が必要な場合は、マイクロプレートリーダー21で590nmと450nmのマイクロプレートを読み取ります。

図3:ブラッドフォードタンパク質アッセイの結果をキャプチャ 。明るい部屋では、マイクロプレートを片手で均一な背景に対してベンチと平行に配置します。一方、スマートフォンはベンチとマイクロプレートに平行に保持されます。 この図の拡大版をご覧になるには、ここをクリックしてください。
5.カラーデータの抽出-自動
- ImageJ と ReadPlate22 のダウンロード、ImageJ プラグインは https://imagej.nih.gov/ij/plugins/readplate/index.html (これは .txt ファイルです) から入手できます。
- ImageJ を開き、[プラグイン] > [インストール ] をクリックして、手順 6.1 でダウンロードしたファイルを選択します。
- [分析]>[測定値の設定]をクリックして測定パラメータを設定し、次のオプションをオンにします。標準偏差;最小および最大のグレー値。平均グレー値。モーダルグレー値。ウィンドウの下部で、[リダイレクト] を [なし] と [小数点以下の桁数 (0-9): 3] に設定します。
- [ファイル]>[開く]に移動し、手順4で撮影したマイクロプレートの写真を選択します。
- ReadPlate>プラグインに移動します。指示を読み、[OK] をクリックします。
- ウェルの数を 96 から選択します。
- プラグインによって自動的にロードされる 矩形選択 ツールを使用して、A1ウェルの中心から始まり、H12ウェルの中心で終わる長方形を作成します。次に、「 OK」をクリックします。
- 青チャンネルを選択し、「OK」をクリックします。
- [ OK]をクリックして、デフォルトのパラメータを確認します。
- ソフトウェアが各ウェル内の領域を描写しているかどうか、および選択した領域が異常な影や反射で領域を覆っていないかどうかを確認します。[ OK] をクリックします。
- 結果を保存し、緑チャンネルに対して手順5.8〜5.10を繰り返します。
- 青と緑の比率は、各色のモードを使用して計算します。
注意: 計算は、手動で行うことも、R、Microsoft Excel、GraphPad Prismなどのリーダー設定のソフトウェアを使用して行うこともできます。
6.カラーデータの抽出-手動
- 無料のオープンソースグラフィックエディタであるInkscapeをダウンロードしてください。
注意: カラーピッカーツール(通常はスポイトとして描かれています)を備えたソフトウェアを使用して、色を識別し、RGBデータを抽出できます。 - Inkscapeを開き、[ファイル]>[開く]に移動します。手順4で撮影したマイクロプレートの画像を選択します。
- 左上に矢印で表示されている オブジェクトの選択と変換(S) ツールを選択し、画像をクリックします。破線の境界線は、選択を示します。
- 左側にスポイトとして描かれている 画像から色を選択(D) ツールを選択します。
- ウェルの中央をクリックします。それに応じて、下部パネルの色(「塗りつぶし:」)が変わります。色をクリックすると、右側に [塗りつぶしとストローク ]タブがポップアップ表示されます。
- [ フラットカラー] ドロップダウンメニューを [RGB]に変更します。各ウェルの青と緑のチャンネルに表示された値を記録します。
- 記録された値を使用して、青と緑の比率を計算します。
注意: 計算は、手動で行うことも、R、Microsoft Excel、GraphPad Prismなどのリーダー設定のソフトウェアを使用して行うこともできます。
7. 検量線の作成と未知数の外挿
- 緑と青の平均強度比をタンパク質標準濃度の関数としてプロットします。
注: データは手動で印刷するか、R、Microsoft Excel、GraphPad Prism などのリーダー設定のソフトウェアを使用してプロットします。 - 各サンプルと希釈液の平均緑と青の強度比を計算します。
- サンプルで得られたシグナルがタンパク質標準試料の線形範囲内にあるかどうかを確認します。
- 検量線の最小値または最大値を下回る値または上回る値は無視します。
- 検量線を表す線形方程式を使用して、サンプル中のタンパク質の量を推定します。それに応じて、計算値に希釈係数を掛けます。
結果
図4は、カラーデータを抽出したマイクロプレートの写真で、450nmと590nmの吸光度を記録しました。ここで代表として報告されたRGBカラーデータは、セクション5で説明したように自動的に取得されました。カラーデータの典型的なパターンは、青の値が増加し、赤と緑の値が減少することです(図5)。すべてのウェルで明らかな反射があり、マイクロ?...
ディスカッション
この論文では、スマートフォンのカメラを使用してブラッドフォードタンパク質アッセイからのデータを記録し、カラーデータを抽出し、生物学的サンプル中のタンパク質レベルを正確に定量する方法であるRGBradfordについて説明します。元のRGBradfordメソッドとの違いの1つは、ここではImageJプラグイン22 でカラーデータを自動的に取得する手順が用いられ?...
開示事項
著者は、宣言すべき利益相反を持っていません。
謝辞
この研究は、ブラジルの国家科学技術開発評議会(CNPq)[助成金番号428048/2018-8および402556/2022-4]とブラジリア大学(ブラジル)から資金提供を受けました。著者は、この研究で使用したスマートフォンへのアクセスを提供してくれたDuarte Nuno Carvalho博士とEvelyn Santos博士(i3s、ポルトガル、ポルト)に感謝します。
資料
| Name | Company | Catalog Number | Comments |
| 96-well flat-bottom polystyrene microtiter plates | Jet Biofil, Guangzhou, China | TCP011096 | Any flat-bottom microplate compativle with optical reading will suffice. |
| Bovine serum albumin | Sigma-Aldrich, St. Louis, MO | A2153 | |
| Coomassie Brilliant Blue G | Sigma-Aldrich, St. Louis, MO | B0770 | |
| Ethyl alcohol | |||
| iPhone 11 | Apple | MWM02BR/A | Can be substituted with other smartphone equiped with a camera |
| iPhone 14 Pro | Apple | N/A | |
| Phosphoric acid | Sigma-Aldrich, St. Louis, MO | 695017 | |
| Redmi Note 9 Pro | XIAOMI | N/A | |
| S22 Ultra | Samsung | N/A | |
| SpectraMax 384 Plus. Microplate reader. | Molecular Devices, San Jose, CA | PLUS 384 | Any microplate reader capable of reading at 450 nm and 590 nm will work. This is optional. The method was actually created to dismiss the need of a microplate reader. |
参考文献
- Zaguri, M., Kandel, S., Rinehart, S. A., Torsekar, V. R., Hawlena, D. Protein quantification in ecological studies: A literature review and empirical comparisons of standard methodologies. Methods in Ecology and Evolution. 12 (7), 1240-1251 (2021).
- Koga, T., et al. Mild electrical stimulation and heat shock ameliorates progressive proteinuria and renal inflammation in mouse model of Alport syndrome. PLoS One. 7 (8), e43852 (2012).
- Peterson, G. L. Determination of total protein. Methods in Enzymology. 91, 95-119 (1983).
- Goldfarb, A. R., Saidel, L. J., Mosovich, E. The ultraviolet absorption spectra of proteins. The Journal of Biological Chemistry. 193 (1), 397-404 (1951).
- Lowry, O. H., Rosebrough, N. J., Farr, A. L., Randall, R. J. Protein measurement with the Folin phenol reagent. The Journal of Biological Chemistry. 193 (1), 265-275 (1951).
- Smith, P. K., et al. Measurement of protein using bicinchoninic acid. Analytical Biochemistry. 150 (1), 76-85 (1985).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 72 (1-2), 248-254 (1976).
- Datki, Z., et al. Application of BisANS fluorescent dye for developing a novel protein assay. PLoS One. 14 (4), e0215863 (2019).
- Van Noorden, R., Maher, B., Nuzzo, R. The top 100 papers. Nature. 514 (7524), 550-553 (2014).
- . Scopus Available from: https://www.scopus.com/ (2022)
- Compton, S. J., Jones, C. G. Mechanism of dye response and interference in the Bradford protein assay. Analytical Biochemistry. 151 (2), 369-374 (1985).
- Chial, H. J., Thompson, H. B., Splittgerber, A. G. A spectral study of the charge forms of Coomassie Blue G. Analytical Biochemistry. 209 (2), 258-266 (1993).
- Pande, S. V., Murthy, M. S. R. A modified micro-Bradford procedure for elimination of interference from sodium dodecyl sulfate, other detergents, and lipids. Analytical Biochemistry. 220 (2), 424-426 (1994).
- Gogstad, G. O., Krutnes, M. -. B. Measurement of protein in cell suspensions using the Commassie brilliant blue dye-binding assay. Analytical Biochemistry. 126 (2), 355-359 (1982).
- Friedenauer, S., Berlet, H. H. Sensitivity and variability of the Bradford protein assay in the presence of detergents. Analytical Biochemistry. 178 (2), 263-268 (1989).
- Stoscheck, C. M. Increased uniformity in the response of the Coomassie blue G protein assay to different proteins. Analytical Biochemistry. 184 (1), 111-116 (1990).
- Zor, T., Selinger, Z. Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Analytical Biochemistry. 236 (2), 302-308 (1996).
- Gee, C. T., Kehoe, E., Pomerantz, W. C. K., Penn, R. L. Quantifying protein concentrations using smartphone colorimetry: A new method for an established test. Journal of Chemical Education. 94 (7), 941-945 (2017).
- de Camargo, C., Vicentini, M., Gobbi, A., Martinez, D., Lima, R. Smartphone for point-of-care quantification of protein by Bradford assay. Journal of the Brazilian Chemical Society. 28 (4), 689-693 (2016).
- Moreira, D. C. RGBradford: Accurate measurement of protein concentration using a smartphone camera and the blue to green intensity ratio. Analytical Biochemistry. 655, 114839 (2022).
- Ernst, O., Zor, T. Linearization of the Bradford Protein Assay. Journal of Visualized Experiments. (38), 1918 (2010).
- Angelani, C. R., et al. A metabolic control analysis approach to introduce the study of systems in biochemistry: the glycolytic pathway in the red blood cell: Metabolic control analysis and the glycolytic pathway. Biochemistry and Molecular Biology Education. 46 (5), 502-515 (2018).
転載および許可
このJoVE論文のテキスト又は図を再利用するための許可を申請します
許可を申請さらに記事を探す
This article has been published
Video Coming Soon
Copyright © 2023 MyJoVE Corporation. All rights reserved