Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Temporal Tracking of Cell Cycle Progression Using Flow Cytometry without the Need for Synchronization
W tym Artykule
Podsumowanie
This protocol describes the use of bromodeoxyuridine (BrdU) uptake to permit the temporal tracking of cells that were in S phase at a specific point in time. Addition of DNA dyes and antibody labeling facilitates detailed analysis of the fate of the S phase cells at later times.
Streszczenie
This protocol describes a method to permit the tracking of cells through the cell cycle without requiring the cells to be synchronized. Achieving cell synchronization can be difficult for many cell systems. Standard practice is to block cell cycle progression at a specific stage and then release the accumulated cells producing a wave of cells progressing through the cycle in unison. However, some cell types find this block toxic resulting in abnormal cell cycling, or even mass death. Bromodeoxyuridine (BrdU) uptake can be used to track the cell cycle stage of individual cells. Cells incorporate this synthetic thymidine analog, while synthesizing new DNA during S phase. By providing BrdU for a brief period it is possible to mark a pool of cells that were in S phase while the BrdU was present. These cells can then be tracked through the remainder of the cell cycle and into the next round of replication, permitting the duration of the cell cycle phases to be determined without the need to induce a potentially toxic cell cycle block. It is also possible to determine and correlate the expression of both internal and external proteins during subsequent stages of the cell cycle. These can be used to further refine the assignment of cell cycle stage or assess effects on other cellular functions such as checkpoint activation or cell death.
Wprowadzenie
The assessment of cell cycle features and changes that occur in cells during cell cycle progression is fundamental to understanding many aspects of biology, particularly cancer biology. Many agents in development for the treatment of malignancies have profound effects on cell cycle progression or induce cell death via cell cycle dependent-mechanisms. In order to study cell cycle dynamics or cells in a particular phase of the cell cycle, it is usual to synchronize cells. However synchronization methods can have detrimental effects on the cells being studied, potentially confounding the results obtained.1 Recently the use of fluorescently tagged proteins that are only present at particular phases of the cells cycle have permitted analysis of cell cycle progression in single cells over time2, however the cells to be studied need to be genetically manipulated to express these tagged proteins, limiting their use to systems where this can be readily achieved.
The cell cycle consists of two active phases: the synthesis (S) phase, where DNA is replicated and mitosis (M) where cell division takes place. These phases are separated by three gap phases, G0, G1 and G2. G0 or quiescence, is a resting phase where the cell has left the cycle, G1 is where the cells increase in size prior to DNA replication and G2 where cell growth continues between completion of DNA replication but before cell division. The progression through the cell cycle is controlled by a number of checkpoints. The G1 checkpoint is activated when environmental conditions are not supportive of DNA synthesis and prevents entry into S phase. The intra-S phase checkpoint or delay can be triggered by DNA damage that may result in stalled replication forks. During G2 the fidelity of the replicated DNA is confirmed and if damage is detected then the G2 checkpoint is activated permitting DNA repair prior to cell division. A final checkpoint during mitosis ensures that chromatids have been correctly aligned at the mitotic plate so that cell division can be successfully completed.3 Activation of these checkpoints is commonly used to synchronize cell populations. Cell cycle checkpoints can be activated by a number of factors but in cancer biology the most common is detection of DNA damage. The DNA damage response is initiated by the PI3-kinase-like kinases ataxia telangiectasia and Rad3 related (ATR) and ataxia telangiectasia mutated (ATM) that activate the downstream effector kinases Chk1 and Chk2, respectively.3 A range of events activates Chk1 including stalled replication forks, DNA crosslinks, and ultraviolet radiation damage while Chk2 is primarily activated by double-strand breaks.
The usual method for studying the effect of altered conditions on the length of the cell cycle is to synchronize the cells in a particular phase of the cell cycle.1 This can be achieved via several methods. Cells can be physically separated based on size, density, side scatter (granularity), and cell surface expression markers. More practically, cells may be synchronized by chemical means. Several agents such as thymidine, hydroxyurea and cytosine arabinoside can be used to inhibit DNA synthesis in the S phase of cell cycle resulting in an accumulation of cells in S phase which continue cycling after the agents are removed. Cells treated with nocodazole, which prevents the formation of the mitotic spindle, arrest with a G2- or M-phase DNA content. Elimination of serum from the culture medium results in the accumulation of cells at G0 phase. The re-addition of the nutrients within the culture serum re-starts the normal cycling of the cells. However, all of these synchronization methods interfere with normal cycling and growth of cells and can result in significant cell death.
Synchronization of acute lymphoblastic leukemia cells is particularly challenging and these cells are not amenable to genetic manipulation. The method described here permits the assessment of cell cycle dynamics and the study of cells in particular phases of the cell cycle without traditional synchronization or genetic modification. This method may also be useful for other cell types where genetic modification and traditional synchronization procedures are not readily achieved. The method is based on the long established use of bromodeoxyuridine (BrdU) incorporation, which has very little impact on the short-term growth and proliferation of cells.4 Established BrdU protocols take advantage of the incorporation of BrdU into newly synthesized DNA during S phase. This permanently marks cells as having been in S phase during BrdU exposure. This population can be identified at later time points by staining for BrdU incorporation and thereby act as a synchronized population that can be followed and assessed over time permitting the study of drug effects on cell cycle transit. BrdU needs to be exposed prior to antibody staining, usually achieved following DNase or acid treatment.6,7 Using flow cytometry to detect incorporated BrdU enables the inclusion of additional markers. The most important is the use of dyes to measure DNA content, enabling the assessment of cell cycle phase distribution of the cells that were in S phase at the start of the study.8 Furthermore additional surface or intracellular antigens can also be studied.9 These may relate to cell cycle events such as Ki67 or to seemingly unrelated cell features such as apoptosis markers like cleaved caspase-3. The potential applications are limited by the imagination of the investigator.
Protokół
The protocol described here uses the acute lymphoblastic leukemia cell line NALM6 but can be applied to other cell types.
1. Solutions and Reagents
- Complete RPMI
- Add 56 ml fetal calf serum (FCS) and 5.5 ml of 200 mM L-glutamine to a 500 ml bottle of RPMI-1640 medium.
- BrdU Stock Solution
- Prepare 32.5 mM BrdU (10 mg/ml) in Dulbecco's Phosphate Buffered Saline (DPBS).
- BrdU Complete RPMI
- Add 6.2 µl of BrdU stock solution to 10 ml of Complete RPMI.
- DNase Solution
- Prepare 1 mg DNase/ml in DPBS.
- Staining Buffer
- Prepare 3% heat-inactivated FCS and 0.09% sodium azide in DPBS.
- Refer to Materials List for definitions of Fixation Buffer, Permeabilization Buffer and Wash Buffer.
2. Cells
Note: Cells were not cultured for greater than 6 months. This method is directly adaptable to any non-adherent cell line with adjustments to cell density and culture media. Use cells that are growing exponentially at the initiation of the experiment.
- Maintain NALM6 cells in T-75 culture flasks in Complete RPMI. Perform all steps under sterile conditions using a Class II Biosafety Cabinet.
- Maintain NALM6 cells between 1-2 x 106 cells per ml by splitting the culture thrice weekly.
- Incubate at 37 °C in 5% CO2 in air.
3. Pulse Labeling of Cells with BrdU
CAUTION: Handle BrdU with care as it is a potential mutagen and teratogen.
- Centrifuge cells at 150 x g for 5 min. Note: Transferring cells into fresh media improves the reproducibility of the results.
- Perform a cell count and resuspend cells in Complete RPMI at 2 x 106 cells/ml.
- Dilute cells 1 in 2 with BrdU Complete RPMI producing a final cell concentration of 1 x 106 cells/ml.
- Incubate at 37 °C with 5% CO2 for 45 min, then dilute cells 1 in 10 with complete RPMI. Centrifuge cells at 150 x g for 5 min and carefully discard all of the supernatant.
- Resuspend cells in a small volume (~100 µl) of complete RPMI, perform a cell count and adjust to 1 x 106 cells/ml.
- Pipette 1 ml of cells into the wells of a 48 well plate. Pipette 1 ml of DPBS into any unoccupied wells to obtain more reproducible results.
- Incubate at 37 °C in 5% CO2 in air for desired timepoints, here 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, and 24 hr. Note: The length of time will depend on what the experimental design aims to measure.
- Transfer all the cells into FACS tubes using a pipette. Rinse the well sequentially with 1 ml volumes of PBS to a final total volume of 5 ml.
- Centrifuge at 150 x g for 5 min and carefully remove all the supernatant. Cells are ready for staining, perform this (section 4) immediately.
4. Cell Staining
Note: If surface staining of cells is required perform it prior to fixation, ensuring that the cells are kept at 4 °C throughout.
- Resuspend cells in 100 µl of staining buffer (for optional surface staining, add the recommended volume of antibody to surface antigens and incubate for 30 min at 4 °C).
- Add 1 ml of staining buffer, centrifuge for 5 min at 150 x g and discard the supernatant.
Note: Specific antibody, concentration, incubation time etc. will vary depending on specific experimental goals. - Fixation and Permeabilization
- Resuspend cells in 100 µl of fixation buffer and incubate for 15 min at room temperature.
- Add 1 ml of wash buffer, centrifuge for 5 min at 150 x g and discard the supernatant.
- Resuspend cells in 100 µl of permeabilization buffer and incubate the cells for 10 min on ice.
- Add 1 ml of wash buffer, centrifuge for 5 min at 150 x g, and discard the supernatant.
- Resuspend cells in 100 µl of fixation buffer per tube and incubate for 5 min at room temperature.
- Add 1 ml of wash buffer, centrifuge for 5 min at 150 x g, and discard the supernatant.
Note: The protocol can be paused here if required. The fixed cells are stable for several days at 4 °C if resuspended in staining buffer. Remove the staining buffer following centrifugation before proceeding.
- DNase Treatment
- Resuspend cells in 100 µl of DNase solution (30 µg of DNase/106 cells) and incubate cells for 1 hr at 37 °C.
- Add 1 ml of wash buffer, centrifuge at 150 x g for 5 min and discard supernatant.
- Antibody Staining
Note: Staining for intracellular markers other than BrdU can be performed simultaneously with the BrdU staining.- IMPORTANT: Prepare compensation controls consisting of unstained cells and cells labeled with each single fluorochrome. Ideally, use the same antibodies for compensation controls as those used in the experimental tubes. However, if this is not feasible, substitute antibodies to highly expressed antigens conjugated to the same fluorochrome.
- Resuspend the cells in 50 µl of wash buffer and add 1 µl/106 cells of BrdU antibody. Note: Directly conjugated antibodies to other specific intracellular antigens can also be added.
NOTE: Antibodies to histone H3 phosphorylated on Ser10 can be used to discriminate between cells in G2 and M, histone H3 is phosphorylated on Ser10 during mitosis.10 Antibodies to cdc2 phosphorylated on Tyr15 can be used to detect cells that have committed to mitosis.11 - Incubate the cells for 20 min at room temperature.
- Add 1 ml of wash buffer, centrifuge cells at 150 x g for 5 min and discard supernatant.
- Stain DNA for Cell Cycle Analysis
- Loosen pellet and add 20 µl of the 7-AAD solution (0.25 µg). Note: It is critical to use a constant amount of 7-AAD/cell.
- Resuspend the cells in 1 ml of Staining buffer.
5. Collection of Flow Cytometry Data
Note: The machine required will depend on the number and nature of the fluorochromes used.
- Collect the following parameters: FSC-A, SSC-A, FSC-H (FSC-W can be used instead of FSC-H) and 7-AAD fluorescence on a linear scale. Collect the APC channel on a log scale. Collect any additional channels required for the assessment of surface or internal labels using a log scale.
- Perform compensation of overlapping signals in emission spectra observed between different fluorochromes before analyzing the samples. Note: Most flow cytometers will perform this automatically.
- Collect at least 10,000 events for each sample.
6. Analysis of Flow Cytometry Data
Note: FlowJo was used in this study for flow cytometry data analysis but other software packages can also be used. The gating strategy is illustrated in Figure 1.
- Identify the viable cell population using FSC-A and SSC-A parameters.
- Within this population exclude doublets and aggregates using FSC-A and FSC-H (FSC-W can also be used here).
- Within this population set a dot plot using 7-AAD on the x-axis and BrdU-APC on the y-axis.
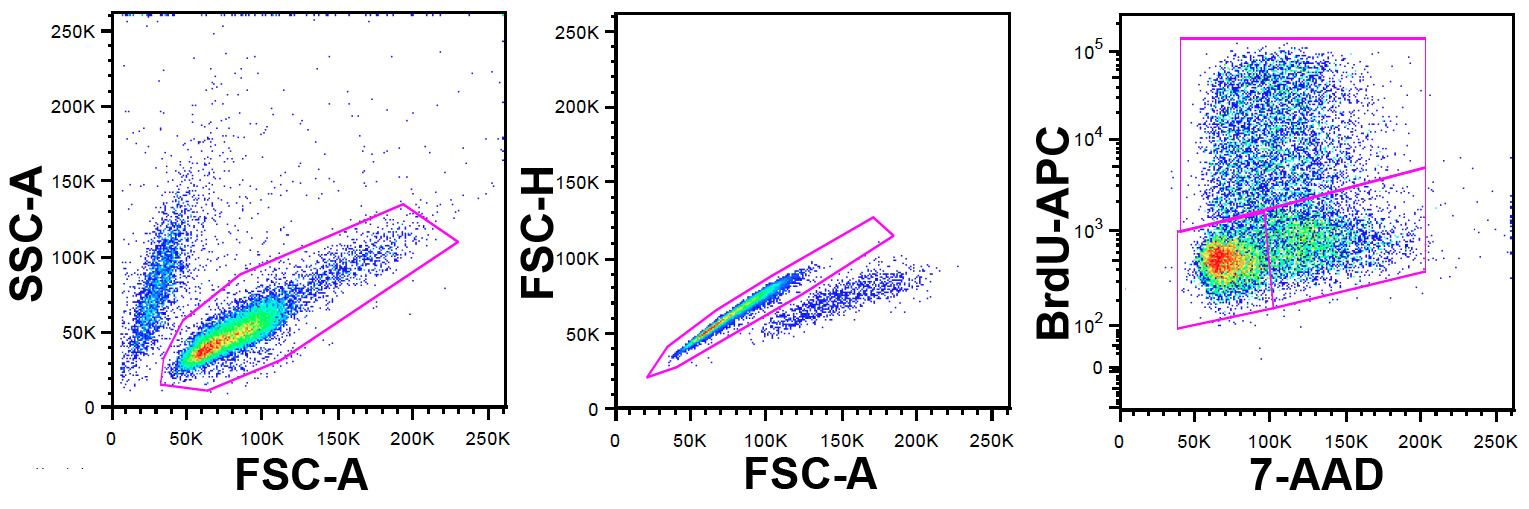
Figure 1: Gating Strategy. Left panel: ungated cells are shown on a FCS-A vs. SSC-A dot plot. The viable cell population is identified by the gate shown. Center panel: cells gated from the left panel are shown on a FSC-A vs. FSC-H dot plot (FSC-W can be used instead of height). Doublets and aggregates are identified, and excluded by the gate shown. Right panel: cells gated from the doublet exclusion date in the center panel are shown on a 7-AAD vs. APC-A dot plot. The BrdU antibody is labeled with APC permitting the identification of cells that have incorporated BrdU during the pulse labeling. 7-AAD provides information on DNA content. The upper gate defines cells positive for BrdU and therefore in S phase at the time of the BrdU pulse, the lower left gate, cells in G0/1 and the lower right gate those in G2/M. Please click here to view a larger version of this figure.
- Cell Cycle Analysis
- Open the first data file and gate on the cells in the doublet exclusion gate.
- Analyze this population for cell cycle distribution (located under platforms in FloJo software) and use the Dean-Jett-Fox model.
- Obtain the positions of the G0/1 and G2/M peaks using create gates.
- Gate on the BrdU positive cells and subject this population to same cell cycle analysis.
- Provide the positions for the G0/1 and G2/M peaks by applying the same gates from create gates and setting constraints (using the created gates) for the positions of the G0/1 and G2/M peaks. This is illustrated in the first 2 panels of Figure 2.
Note: Other software may also be used to analyze the data and the instructions would vary accordingly.

Figure 2: Cell Cycle Progression. The first panel (All Cells) is gated on the cell population defined by the doublet exclusion gate. This population was displayed in a histogram with 7-AAD on the X-axis. The peak of the G0/1 peak is indicated by the arrow below the axis. In subsequent panels BrdU positive cells have been gated on as shown in Figure 1. The value for the G0/1 position obtained when gating of the doublet exclusion gate is applied to the BrdU positive gated cells within FlowJo cell cycle software. Each subsequent panel was gated on the BrdU positive population as shown in Figure 1 and the position of the G0/1 peak based on the value obtained when analyzing the whole population as shown in the first two panels. Using the BrdU negative fraction to identify the location of the G0/1 population for the BrdU positive cells in the same sample controls for any slight differences in the intensity of the DNA stain between samples. The number shown on each panel represents the time since the BrdU pulse ended. The calculated cell cycle phases are shown in shaded green. Please click here to view a larger version of this figure.
Wyniki
This methodology can be used to obtain a range of information. A few applications are outlined here.
Assessment of the duration of the cell cycle
To determine the time required for cells to transit through the cell cycle, cells are harvested at various time points following the BrdU pulse. The intervals between assessments can be adapted to the particular cells being analyzed. Hematopoietic cell lines were assessed every hour over a 24 hr period in the absence of an...
Dyskusje
The ability to analyze the cell cycle is important for the understanding of cancer biology and the mechanism of action of both drugs and genes that influence cell proliferation and growth. While there are a multitude of assays that reportedly measure cell proliferation, the majority only provide a measure that indicates the number cells present. These include assays that measure cell number by direct visualization and counting, metabolic activity or ATP concentration. The main advantage of many of these methods is that t...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
The work was funded by the Leukemia and Lymphoma Society of the USA (6105-08), a Cancer Council NSW grant (13-02), an NHMRC Senior Research Fellowship (LJB) (1042305) and project grant (1041614).
Materiały
| Name | Company | Catalog Number | Comments |
| APC BrdU Flow Kit | BD Biosciences | 552598 | Contains BrdU antibody, 7-AAD and BD Cytofix/Cytoperm Buffer (referred to as Fixation Buffer) |
| BD Cytoperm Permeabilization Buffer Plus | BD Biosciences | 561651 | Referred to as Permeabilization buffer |
| BD Perm/Wash Buffer | BD Biosciences | 554723 | Referred to as Wash buffer |
| DNase | Sigma | D-4513 | |
| BD Falcon 12 x 75 mm FACS tubes | BD Biosciences | 352008 | |
| BD Pharmingen Stain Buffer | BD Biosciences | 554656 | |
| BD LSR FORTESSA flow cytometer | BD Biosciences | FORTESSA | |
| Pipetman | Gilson | P2, P20, P100, P1000 | |
| RPMI 1,640 w/o L-Gln 500 ml | Lonza | 12-167F | |
| DPBS | Lonza | 17-512F | |
| Fetal Bovine Serum | FisherBiotec | FBS-7100113 | |
| L-Glutamine | Sigma | G7513-100ML | |
| 5-Bromo-2′-deoxyuridine | Sigma | B5002-1G | |
| Falcon TC 150 cm2 vented Flasks | BD Biosciences | 355001 | |
| Pipettes 25 ml | Greiner | 760180 | |
| Aersol Pipettes 200 µl | Interpath | 24700 | |
| Aersol Pipettes 1 ml | Interpath | 24800 | |
| Centrifuge | Spintron | GT-175R | |
| CO2 incubator | Binder | C 150 | |
| AF488 anti-Histone H3 Phospho (Ser10) Antibody | Cell Signalling | 9708S | |
| Phospho-Chk2 (Thr68) (C13C1) Rabbit mAb | Cell Signalling | 2197S | |
| Phospho-Chk1 (Ser345) (133D3) Rabbit mAb | Cell Signalling | 2348S | |
| NALM6 | DSMZ | ACC-128 |
Odniesienia
- Banfalvi, G., Banfalvi, G. . Methods Mol Biol. 761, 1-23 (2011).
- Sakaue-Sawano, A., et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 132, 487-498 (2008).
- Harper, J. W., Elledge, S. J. The DNA damage response: ten years after. Molecular Cell. 28, 739-745 (2007).
- Latt, S. A., George, Y. S., Gray, J. W. Flow cytometric analysis of bromodeoxyuridine-substituted cells stained with 33258 Hoechst. The Journal of Histochemistry and Cytochemistry. 25, 927-934 (1977).
- Gratzner, H. G. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science. 218, 474-475 (1982).
- Carayon, P., Bord, A. Identification of DNA-replicating lymphocyte subsets using a new method to label the bromo-deoxyuridine incorporated into the DNA. Journal of Immunological Methods. 147, 225-230 (1992).
- Gonchoroff, N. J., et al. S-phase detection with an antibody to bromodeoxyuridine. Role of DNase pretreatment. Journal of Immunological Methods. 93, 97-101 (1986).
- Rabinovitch, P. S., Torres, R. M., Engel, D. Simultaneous cell cycle analysis and two-color surface immunofluorescence using 7-amino-actinomycin D and single laser excitation: applications to study of cell activation and the cell cycle of murine Ly-1 B cells. Journal of Immunology. 136, 2769-2775 (1986).
- Saunders, P. O., et al. RAD001 (everolimus) induces dose-dependent changes to cell cycle regulation and modifies the cell cycle response to vincristine. Oncogene. 32, 4789-4797 (2013).
- Hendzel, M. J., et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106, 348-360 (1997).
- Draetta, G., Beach, D. Activation of cdc2 protein kinase during mitosis in human cells: cell cycle-dependent phosphorylation and subunit rearrangement. Cell. 54, 17-26 (1988).
- Saunders, P. O., et al. RAD001 (everolimus) induces dose-dependent changes to cell cycle regulation and modifies the cell cycle response to vincristine. Oncogene. , (2012).
- Knudsen, R. C., Ahmed, A. A., Sell, K. W. An in vitro microassay for lymphotoxin using microculture plates and the multiple automated sample harvester. Journal of Immunological Methods. 5, 55-63 (1974).
- Beck, H. P. Proliferation kinetics of perturbed cell populations determined by the bromodeoxyuridine-33258 technique: radiotoxic effects of incorporated [3H]thymidine. Cytometry. 2, 170-174 (1981).
- Miller, M. J., Wei, S. H., Parker, I., Cahalan, M. D. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 296, 1869-1873 (2002).
- Collins, J. M., Berry, D. E., Bagwell, C. B. Different rates of DNA synthesis during the S phase of log phase HeLa S3, WI-38, and 2RA cells. Journal of Biological Chemistry. 255, 3585-3590 (1980).
- Schwarting, R., Gerdes, J., Niehus, J., Jaeschke, L., Stein, H. Determination of the growth fraction in cell suspensions by flow cytometry using the monoclonal antibody Ki-67. Journal of Immunological Methods. 90, 65-70 (1986).
- Danova, M., et al. Cell cycle-related proteins: a flow cytofluorometric study in human tumors. Biology of the Cell. 64, 23-28 (1988).
- Juan, G., et al. Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry. 32, 71-77 (1998).
- Langan, T. J., Chou, R. C. Synchronization of mammalian cell cultures by serum deprivation. Methods in Molecular Biology. 761, 75-83 (2011).
- Kues, W. A., et al. Cell cycle synchronization of porcine fetal fibroblasts: effects of serum deprivation and reversible cell cycle inhibitors. Biology of Reproduction. 62, 412-419 (2000).
- Urbani, L., Sherwood, S. W., Schimke, R. T. Dissociation of nuclear and cytoplasmic cell cycle progression by drugs employed in cell synchronization. Experimental Cell Research. 219, 159-168 (1995).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone