Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Visualizing Diffusional Dynamics of Gold Nanorods on Cell Membrane using Single Nanoparticle Darkfield Microscopy
W tym Artykule
Podsumowanie
Here, we show the use of traditional dark-field microscopy to monitor the dynamics of gold nanorods (AuNRs) on cell membrane. The location and orientation of single AuNRs are detected using ImageJ and MATLAB, and the diffusive states of AuNRs are characterized by single particle tracking analysis.
Streszczenie
Analyzing the diffusional dynamics of nanoparticles on cell membrane plays a significant role in better understanding the cellular uptake process and provides a theoretical basis for the rational design of nano-medicine delivery. Single particle tracking (SPT) analysis could probe the position and orientation of individual nanoparticles on cell membrane, and reveal their translational and rotational states. Here, we show how to use traditional dark-field microscopy to monitor the dynamics of gold nanorods (AuNRs) on live cell membrane. We also show how to extract the location and orientation of AuNRs using ImageJ and MATLAB, and how to characterize the diffusive states of AuNRs. Statistical analysis of hundreds of particles show that single AuNRs perform Brownian motion on the surface of U87 MG cell membrane. However, individual long trajectory analysis shows that AuNRs have two distinctly different types of motion states on the membrane, namely long-range transport and limited-area confinement. Our SPT methods can be potentially used to study the surface or intracellular particle diffusion in different biological cells and can become a powerful tool for investigations of complex cellular mechanisms.
Wprowadzenie
The dynamics of nanoparticles (NPs) on the membrane is closely associated with the cellular uptake process, which is essential for the understanding of cell functions, viral or bacterial infections and the development of artificial nanomedical delivery systems1,2. Single particle tracking (SPT) technique is a robust tool for characterizing the heterogeneous behaviors of NPs3,4. In general, cell membrane is fluidic, which means that the components such as proteins and lipids can move laterally in the plasma membrane plane5,6,7. The spatiotemporal complexity of membrane organization and structure may lead to spatiotemporal heterogeneity of the interaction between NPs and membrane. Hence, direct visualization of the movement of NPs on the membrane requires both high spatial and temporal resolution.
Single particle tracking microscopy that monitors the localization of individual particles in living cells with a spatial resolution of tens of nanometers and a time resolution of milliseconds has been well developed to study the NPs or membrane molecules dynamics8,9. Fluorescence-based microscopic imaging techniques have become valuable tools for observing NPs/molecules in living cell environment9,10,11,12. For example, total internal reflection fluorescence microscopy, which images thin layers (~100 nm) of the sample at the substrate/solution interface with a high spatiotemporal resolution has been widely used in studies of membrane molecules dynamics13,14. However, the inherent disadvantages of single fluorophores, such as low intensity and rapid irreversible photobleaching reduce the accuracy and duration of tracking13. Therefore, non-fluorescent plasmonic NPs, which replace the fluorescent probes, have attracted more and more attention in long-term imaging studies due to their unique optical characteristics15. Based on the scattering signals of plasmonic NP probes, several kinds of optical microscopic imaging technologies have been used to study the mechanism of biological processes, such as dark-field microscopy (DFM)16, interferometric scattering (iSCAT) microscopy17 and differential interference contrast microscopy (DICM)18. In addition, the motion and rotation dynamic of AuNRs can be obtained using DFM and DICM18,19,20,21,22. Typically, in an SPT experiment, the motion of the object is recorded by the optical microscope, and then analyzed by SPT analysis methods3. The time-resolved trajectories and orientational angles generated by individual NPs are normally stochastic and heterogeneous, so it is necessary to present abundant dynamic information with various analysis methods.
Here, we provide an integrated protocol that monitors the dynamics of AuNRs on cell membrane using DFM, extracts the location and orientation of AuNRs with ImageJ and MATLAB and characterizes the diffusion of AuNRs with SPT analysis methods. As a demonstration, we show here how to use the SPT protocol to visualize dynamics of unmodified AuNRs (CTAB-AuNRs, synthesized by cetyltrimethylammonium ammonium bromide molecule as protective agent) on U87 MG cell membrane. It has been demonstrated that CTAB-AuNRs can adsorb proteins in biological environment, move on cell membrane and then enter cells2,20,22. U87 MG cell is the most common and most malignant tumor of the central nervous system, and its membrane receptors are abnormally expressed. The membrane receptors can interact with proteins on AuNRs, which influence the dynamics of AuNRs. Our protocol is generally applicable to other SPT experiments in the field of biology.
Access restricted. Please log in or start a trial to view this content.
Protokół
1. Cell culture
- Prepare complete medium for U87 MG cells by adding fetal bovine serum (final concentration 10%) and penicillin-streptomycin (final concentration 1%) to the minimum essential medium (MEM). Use plastic cell culture dish for cells subculture.
- Passage cells 2 to 3 times a week.
- Remove the culture medium and rinse the cell layer with Dulbecco's phosphate-buffered saline (D-PBS) 2~3 times when confluent (80%~90%).
- Add 1.0 to 2.0 mL of Trypsin-EDTA solution to the cell culture dish and observe cells under an inverted microscope until cells become round (3~5 min).
- Add 3.0 mL of prepared complete medium and disperse the cells by gently pipetting.
- Add cell suspension (1 mL) to new culture dish with fresh cell medium (3 mL) and resuspend the cells.
- Maintain the cells at 37 °C and 5% CO2 in a humidified atmosphere.
2. Microscope slide preparation
NOTE: U87 MG cells of third to tenth generation with high activity are used in SPT experiments.
- Sterilize 22 mm × 22 mm coverslips already cleaned with Piranha solution by immersing in ethanol (99.9%).
- Use forceps to take out the cover slip from the ethanol solution (step 2.1) and sterilize by burning ethanol on the flame. Once all ethanol is burnt, place coverslips in a plastic cell culture dish (35 x 10 mm) filled with 2 mL of cell medium (no phenol red).
- Add 50 µL of the cell suspension from step 1.2.3 on the coverslip and gently push the dish back and forth and left and right to evenly distribute the cells. Place it in a humidified atmosphere.
- When U87 MG cells on the coverslip reach 20%–40% confluency (~12 h), add 20 μL of CTAB-AuNRs (138 pM) into the dish and disperse. Incubate in humidified atmosphere for 5 min.
- Add 100 μL of the culture medium (no phenol red) from dish in step 2.4 into the groove of the grooved glass slide (Figure 1) which is precleaned with Piranha solution.
- Take the coverslip out from the dish and inverted on the top of the groove of the microscope glass slide (Figure 1). Seal with nail polish, let it dry and place it on the stage to perform SPT experiments.
3. Performing single particle tracking experiments with darkfield microscopy (Figure 1).
- Place a drop of oil on the oil-immersed darkfield condenser (NA 1.43-1.20) and turn the knob to make the condenser contact the glass slide.
- Put a drop of oil on the top of the cover glass and turn the focusing knob to make the 60x oil immersion objective (NA 0.7–1.25) touch the oil.
- Turn on the light source and slightly turn the focusing knob to focus the imaging plane.
NOTE: In the field of view, the background is black, the cells are bright, and the CTAB-AuNRs (aspect ratio~2:1, Figure 2) are small colored (red, yellow, or green) scattering spots. - Capture sample scattering light by a color CMOS camera. Click “Camera icon” in the software to record and export TIFF format to save images.
4. Data acquisition
- Extract single long-term trajectory
- Operate as described in Figure 3 to convert the time-series dark-field images from “RGB color” mode to “8 bit” mode. In the Image J click Image | Type | 8 bit. To adjust the contrast, click Image | Adjust | Brightness | Contrast.
- Select a target particle and cutoff the time-series backgrounds by boxing and deleting the background with "Ctrl+X".
- Open the particle detection and particle Linking window by clicking the “Plugins | Particle Tracker Classic | Particle Tracker”.
- Set Radius to 6, Cutoff to 0 and Percentile to 0.01%.
NOTE: To detect the particle, adjust above three parameters with the assistance of Preview. Ensure that radius is slightly larger than the targeted particle and smaller than the smallest inter-particle separation. Percentile is the lower limit of intensity distribution that to be candidate particles. - Set the Link Range to 5 and Displacement to 10.
NOTE: To link the particle between consecutive adjacent frames, adjust the above two parameters. Displacement is the maximum pixels that a particle can move between two succeeding frames, and Link Range is the number of consecutive frames to consider when determining the best corresponding match. - Click “OK” to open the ParticleTracker Results window to see the results.
- Click “Visualize All Trajectories” to inspect the generated trajectories.
- Click “Relink Particles” menu at the top to re-link the detected particles with different link range and percentile parameters, if the software generated trajectory does not match the moving trajectory of the AuNR.
- Click “Save Full Report” to save results if the software generated trajectory and the moving trajectory of the AuNR are matched.
NOTE: An example of extracting single long-term trajectory with Image J is shown in Figure 4.
- Extracting Multi-trajectories
NOTE: An example of extracting multi-trajectories with Fiji is shown in Figure 5. There are several stages, and each stage constitutes a step in the tracking process. The result of each step is displayed immediately, which allows the user to go back to readjust settings when the output is unsatisfactory.- Operate as described in Figure 3 to convert the time-series dark-field images from “RGB color” mode to “8 bit” mode. In the Image J pathway click “Image | Type | 8 bit”. To adjust their contrast, click “Image | Adjust | Brightness | Contrast”.
- Open the start panel by clicking “Plugins | Tracking | TrackMate”. Click “Next”.
- Choose the “LoG detector” from the drop-down menu and click “Next”.
- Adjust parameters of the detector configuration panel. Set Estimated blob diameter to 10, set Threshold to 0, and Select “Do Sub-pixel Localization”.
- Click “Next” to open initial spot filtering panel.
NOTE: Several parameters can be adjusted to further optimize the target points. In this example, no other parameters were adjusted. - Click “Next” and select “HyperStack displayer” from the drop-down menu.
- Click “Next” to open Spot filtering panel. Set the Quality above 1.88, X above 38.86, Y above 56.54.
- Click “Next” and select “Simple LAP tracker” from the drop-down menu. Configure the simple LAP tracker by adjusting three parameters, that is, the Linking max distance = 15, Gap-closing max distance = 15 and Gap-closing max frame gap = 5.
NOTE: Linking-max distance is the maximum displacement of a point between two frames. Gap-closing max distance is the maximum displacement of two segments. Gap-closing max frame gap is the largest frame between two points to be bridged. - Click “Next” to track. When finished, continue to click “Next”.
- Set filters on tracks, such as set Number of spots in track above 300.
- Keep clicking “Next” until the final save panel open. Select “Export tracks to XML file” from the drop-down menu, and then click “Execute” to save in csv format.
- R/G values
NOTE: Scattering intensities of targeted AuNRs in the R and G channels are obtained from color darkfield images by using a code written in MATLAB (https://github.com/fenggeqd/JOVE-2020/tree/master/RGandPolarangle), and the extraction principle is presented in Figure 6.- Use the function of xycoordination.m to find the center pixel coordinate of the AuNR in each frame according to the x-y coordinate (extracted by ImageJ/Fiji).
- Use the function of RGextraction.m to delimit a 3 x 3 pixels matrix, extract the 9 scattering-intensity values of R or G channels, and calculate an average value (μ, defined as R or G).
NOTE: The 3 x 3 pixels matrix is centered on the pixel coordinates obtained from step 4.3.1.
- Polar angle
NOTE: The polar angle is the angle between the longitude of the AuNR and the optical axis (as shown in Figure 1), which can reflect the spatial (Z axis) rotation dynamics of the AuNR.- Use the function of polarangle.m to calculate the polar angles (θ) by the dual-channel differential method22,
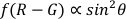 .
.
- Use the function of polarangle.m to calculate the polar angles (θ) by the dual-channel differential method22,
5. Data analysis
NOTE: A systematic and robust data analysis framework is essential for the performance and efficiency of SPT analysis methods. The custom software written in MATLAB is used (https://github.com/fenggeqd/JOVE-2020/tree/master/Analysis_parameters). A graphing and analysis software (see Table of Materials) is used for drawing the plots.
- Analysis parameters
- Use scripts of csv_data_extract_dis_vel_ss.m and csv_data_MSD.m to calculate dynamic parameters according to formulas shown in Table 1.
NOTE: These parameters are used to analyze the dynamics of AuNRs and consist of three parts. (1) Trajectory related parameters: displacement, step size, velocity, radius of gyration (Rg), and turning angle (Ta); (2) MSD parameters: diffusion coefficient (Dt) and abnormal diffusion exponent (α); and (3) Rotation related parameters: polar angle and rotational lability (σ).
- Use scripts of csv_data_extract_dis_vel_ss.m and csv_data_MSD.m to calculate dynamic parameters according to formulas shown in Table 1.
| Measures | Definition | Physical meaning | |
| Displacement | 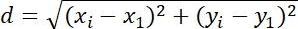 | Changes in position of objects | |
| Step size | 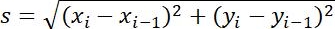 | Distance between two adjacent points | |
| Velocity |  | Speed of objects motion | |
| Rg | 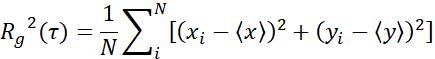 | Moving range of objects in a specific time interval | |
| Ta | 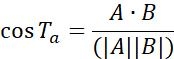 | Motion direction of objects between two adjacent points | |
| MSD | 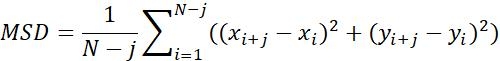 | Average moving distance of objects in a specific time interval | |
| Dt | 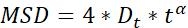 | Diffusion ability of objects | |
| α | 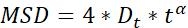 | Normal diffusion (α~1) | |
| Polar angle |  | 3D orientation information of objects | |
| σ |  | Degree of dispersion of the polar angle data set | |
Table 1: Three types of parameters used for analysis. These include trajectory related parameters (displacement, step size, velocity, Rg and Ta), MSD parameters (MSD, Dt and α) and rotation related parameters (polar angle and rotational lability).
- Visual analysis of trajectory
NOTE: Trajectory visualization can intuitively present the spatiotemporal heterogeneity of particle motion and the trajectory (coordinate) distribution of dynamic parameters, such as time-mapping trajectory, Rg- and Dt-mapping trajectory, and polar-angle mapping trajectory. Mapping trajectories were drawn using the graphing and analysis software.- Set x coordinate as X, y coordinate as Y, and time (Rg, Dt, polar angle) as Z.
- Click “Plot | Scatter plot | Color-mapping Plot”.
- Add the color bar.
- MSD analysis
NOTE: Motion activity and motion mode of the particles can be obtained by MSD analysis23. The larger the Dt, the more active the diffusion motion of the particles. when α~1, particles do normal diffusion motion, otherwise they perform abnormal diffusion motion.- MSD-τ figures
- Set time interval (τ) as X, MSD data as Y.
- Click “Plot | Scatter plot”
- Fit data by clicking “Analyze | Fitting | Nonlinear curve fitting (Function: Allometricl)”.
- MSD-τ double-logarithmic figure
NOTE: MSD-τ double-logarithmic figure, whose slope is α and intercept is Dt, can intuitively present particle motion.- Set logarithmic time interval as X, and logarithmic MSD as Y.
- Click “Plot | Scatter plot”.
- Fit data by clicking “Analyze | Fitting | Linear curve fitting”.
- D-τ figure (MSD/4t)
NOTE: D=MSD/4τ, is a function of time τ and anomaly factor α, and the D-τ figure directly shows the change of diffusion coefficient with time. When D increases with time, α is greater than 1 and particles do superdiffusion motion.- Set logarithmic time interval (τ) as X, and logarithmic D as Y.
- Click “Plot | Scatter plot”.
- Fit data by clicking “Analyze | Fitting | Nonlinear curve fitting (Function: “Allometricl”)”.
NOTE: The shorter the trajectories, the higher the inaccuracy of the diffusion estimates. In general, Dt and α through MSD-τ analysis of long interval time (> 30 τ) were obtained. However, simple and rough fitting will smooth out motion details. Hence, MSD-τ analysis of short interval time (< 10 τ) should be performed to analyze the motion behavior of particles in a short time.
- MSD-τ figures
- Statistical analysis
- Multi-particles statistical analysis
NOTE: Multi-particles statistical analysis can reflect the motion state of particles in a spatial region, which indirectly indicates the spatial heterogeneity environment. For instance, if the histogram of Dt exhibits a large-scale distribution or multi-peaks distribution, it means that the motion activities of particles are heterogeneous.- Set dynamic parameters (such as Dt, Rg, max displacement) as Y.
- Click “Plot | Histogram”.
- Double click the histogram, and set division size or the number of divisions. Click “Apply”.
- Single-particle statistical analysis
NOTE: The statistical analysis of single particles can show the motion behavior of individual particles, which also indirectly reflects the spatiotemporal heterogeneity of the surrounding environment.- Multiple frames
- Calculate dynamic parameters (such as Ta, step size, polar angle) of all frames of single long trajectory, and copy to Origin table and set as Y.
- Click “Plot | Histogram”.
- Double click the histogram, and set division size or the number of divisions. Click “Apply”.
NOTE: If both Ta and step size exhibit a small value distribution, the particles do a small-step super-diffusion motion.
- Moving windows
- Calculate dynamic parameters (such as Rg, Dt) of all frames of single long trajectory through moving-window method (11 frames), and copy to Origin table and set as Y.
- Click “Plot | Histogram”.
- Double click the histogram, and set division size or the number of divisions. Click “Apply”.
- Multiple frames
- Multi-particles statistical analysis
- Time-series analysis
NOTE: Statistical analysis can reveal the motion state of NPs, and time-series analysis can present the motion behavior as a supplement. Combining several time-series parameters, it can discriminate the motion behavior of NPs at the temporal and spatial levels.- Set time as X, time-series parameters as Y (such as displacement, polar angle and Rg).
- Click “Plot | Multi-pane diagram | Stacked plot | Line + Symbol”.
Access restricted. Please log in or start a trial to view this content.
Wyniki
In the protocol, the unmodified 40 x 85 nm CTAB-AuNRs were used. As shown in Figure 2B, its longitudinal plasmonic maximum at is ~650 nm (red region) and transverse resonance is at 520 nm (green region). Previous literatures have revealed that the optical properties (such as LSPR intensity) of plasmonic AuNRs will change significantly with their diameter20,22. In Figure 2C, the scattering intensity from ...
Access restricted. Please log in or start a trial to view this content.
Dyskusje
The presented protocol is used to study the dynamics of AuNRs on cell membrane. The protocol consists of four parts, including microscopic imaging, data extraction, dynamic parameters calculation and data analysis methods, and each part is flexible and universal. Therefore, there are many possible future applications, for instance, studying movement of NP-linked membrane molecules on membrane, endocytosis dynamics of NP-labeled receptors, dynamic analysis of intracellular NPs and vesicle-coated NPs transportation along m...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
The authors have nothing to disclose.
Podziękowania
This work was supported by the National Natural Science Foundation of China with grant numbers of 21425519, 91853105 and 21621003.
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| CTAB coated gold nanorods(CTAB-AuNRs) | Nanoseedz | NR-40-650 | 85 nm * 40 nm |
| Color CMOS camera | Olympus | DP74 | Japan |
| Coverslips | Citoglas | z10212222C | 22*22 mm |
| Dark-field microscopy | Nikon | 80i | upright microscope |
| Fetal bovine serum (FBS) | Gibco | 10099141 | |
| Fiji | National Institutes of Health | 2.0.0-rc-69/1.52 p | a distribution of ImageJ |
| Grooved glass slide | Sail brand | 7103 | Single concave |
| Image J | National Institutes of Health | 1.52 j | |
| MATLAB | MathWorks | R2019b | |
| MATLAB Code | https://github.com/fenggeqd/JOVE-2020 | ||
| Minimum essential medium (MEM) | Gibco | 10-010-CVR | with phenol red |
| Minimum essential medium (MEM) | Gibco | 51200038 | no phenol red |
| Origin | OriginLab | Origin Pro 2018C | |
| Penicillin-streptomycin | Gibco | 15140122 | |
| Plastic cell culture dishes | Falcon | 353002 | |
| Plastic cell culture dishes | Falcon | 353001 | 35*10 mm |
| U87 MG cell | American Type Culture Collection | ATCC HTB-14 | a human primary glioblastoma cell line |
Odniesienia
- Rees, P., Wills, J. W., Brown, M. R., Barnes, C. M., Summers, H. D. The origin of heterogeneous nanoparticle uptake by cells. Nature Communication. 10 (1), 2341(2019).
- Behzadi, S., et al. Cellular uptake of nanoparticles: journey inside the cell. Chemical Society Reviews. 46 (14), 4218-4244 (2017).
- Shen, H., et al. Single Particle Tracking: From Theory to Biophysical Applications. Chemical Reviews. 117 (11), 7331-7376 (2017).
- Saxton, M. J. Single-particle tracking: connecting the dots. Nature Methods. 5 (8), 671-672 (2008).
- Kusumi, A., et al. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson's fluid-mosaic model. Annual Review of Cell and Developmental Biology. 28, 215-250 (2012).
- Jacobson, K., Liu, P., Lagerholm, B. C. The Lateral Organization and Mobility of Plasma Membrane Components. Cell. 177 (4), 806-819 (2019).
- Sezgin, E., Levental, I., Mayor, S., Eggeling, C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nature Reviews Molecular Cell Biology. 18 (6), 361-374 (2017).
- von Diezmann, A., Shechtman, Y., Moerner, W. E. Three-Dimensional Localization of Single Molecules for Super-Resolution Imaging and Single-Particle Tracking. Chemical Reviews. 117 (11), 7244-7275 (2017).
- Rosenberg, J., Huang, J. Visualizing Surface T-Cell Receptor Dynamics Four-Dimensionally Using Lattice Light-Sheet Microscopy. Journal of Visualized Experiments. (155), e59914(2020).
- Kusumi, A., Tsunoyama, T. A., Hirosawa, K. M., Kasai, R. S., Fujiwara, T. K. Tracking single molecules at work in living cells. Nature Chemical Biology. 10 (7), 524-532 (2014).
- Rocha, J. M., Gahlmann, A. Single-Molecule Tracking Microscopy - A Tool for Determining the Diffusive States of Cytosolic Molecules. Journal of Visualized Experiments. (151), e59387(2019).
- Uphoff, S., Sherratt, D. J., Kapanidis, A. N. Visualizing protein-DNA interactions in live bacterial cells using photoactivated single-molecule tracking. Journal of Visualized Experiments. (85), e511177(2014).
- Pinaud, F., Clarke, S., Sittner, A., Dahan, M. Probing cellular events, one quantum dot at a time. Nature Methods. 7 (4), 275-285 (2010).
- Mehidi, A., et al. Transient Activations of Rac1 at the Lamellipodium Tip Trigger Membrane Protrusion. Current Biology. 29 (17), 2852-2866 (2019).
- Ye, Z., Wang, X., Xiao, L. Single-Particle Tracking with Scattering-Based Optical Microscopy. Analytical Chemistry. 91 (24), 15327-15334 (2019).
- Pan, Q., Zhao, H., Lin, X., He, Y. Spatiotemporal Heterogeneity of Reactions in Solution Observed with High-Speed Single-Nanorod Rotational Sensing. Angewandte Chemie. International Ed. in English. 58 (25), 8389-8393 (2019).
- Taylor, R. W., et al. Interferometric scattering microscopy reveals microsecond nanoscopic protein motion on a live cell membrane. Nature Photonics. 13 (7), 480-487 (2019).
- Chen, K., et al. Characteristic rotational behaviors of rod-shaped cargo revealed by automated five-dimensional single particle tracking. Nature Communication. 8 (1), 887(2017).
- Xu, D., He, Y., Yeung, E. S. Y. Direct observation of the orientation dynamics of single protein-coated nanoparticles at liquid/solid interfaces. Angewandte Chemie. International Ed. in English. 53 (27), 6951-6955 (2014).
- Lehui, X., Yan, H., Edward, S. Y. Three Dimensional Orientational Imaging of Nanoparticles with Darkfield Microscopy. Analytical Chemistry. 82, 5268-5274 (2010).
- Ge, F., Xue, J., Wang, Z., Xiong, B., He, Y. Real-time observation of dynamic heterogeneity of gold nanorods on plasma membrane with darkfield microscopy. Science China Chemistry. 62, 1072-1081 (2019).
- Tinevez, J. Y., et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115, 80-90 (2017).
- Janczura, J., Weron, A. Ergodicity testing for anomalous diffusion: small sample statistics. The Journal of Chemical Physics. 142 (14), 144103(2015).
- Kim, D. H., et al. Single particle tracking-based reaction progress kinetic analysis reveals a series of molecular mechanisms of cetuximab-induced EGFR processes in a single living cell. Chemical Science. 8 (7), 4823-4832 (2017).
- Kurzthaler, C., et al. Probing the Spatiotemporal Dynamics of Catalytic Janus Particles with Single-Particle Tracking and Differential Dynamic Microscopy. Physical Review Letters. 121 (7), 078001(2018).
- Lin, X., Pan, Q., He, Y. In situ detection of protein corona on single particle by rotational diffusivity. Nanoscale. 11 (39), 18367-18374 (2019).
- Wei, L., et al. Sub-diffraction-limit localization imaging of a plasmonic nanoparticle pair with wavelength-resolved dark-field microscopy. Nanoscale. 9 (25), 8747-8755 (2017).
- Cheng, X., Dai, D., Xu, D., He, Y., Yeung, E. S. Subdiffraction-limited plasmonic imaging with anisotropic metal nanoparticles. Analytical Chemistry. 86 (5), 2303-2307 (2014).
- Belyy, V., et al. PhotoGate microscopy to track single molecules in crowded environments. Nature Communication. 8, 13978(2017).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone