Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Assessment of Intestinal Transcytosis of Neonatal Escherichia coli Bacteremia Isolates
W tym Artykule
Podsumowanie
Escherichia coli causes sepsis in neonates who ingest the bacteria around the time of birth. The process involved in E. coli’s ability to travel from the enteric tract to the bloodstream is poorly understood. This in vitro model assesses the ability of E. coli strains to travel through the intestinal epithelial cells.
Streszczenie
Newborns ingest maternal E. coli strains that colonize their intestinal tract around the time of delivery. E. coli strains with the ability to translocate across the gut invade the newborn's bloodstream, causing life-threatening bacteremia. The methodology presented here utilizes polarized intestinal epithelial cells grown on semipermeable inserts to assess the transcytosis of neonatal E. coli bacteremia isolates in vitro. This method uses the established T84 intestinal cell line that has the ability to grow to confluence and form tight junctions and desmosomes. After reaching confluence, mature T84 monolayers develop transepithelial resistance (TEER), which can be quantified using a voltmeter. The TEER values are inversely correlated with the paracellular permeability of extracellular components, including bacteria, across the intestinal monolayer. The transcellular passage of bacteria (transcytosis), on the other hand, does not necessarily alter the TEER measurements. In this model, bacterial passage across the intestinal monolayer is quantified for up to 6 h post-infection, and repeated measurements of TEER are made to monitor the paracellular permeability. In addition, this method facilitates the use of techniques such as immunostaining to study the structural changes in tight junctions and other cell-to-cell adhesion proteins during bacterial transcytosis across the polarized epithelium. The use of this model contributes to the characterization of the mechanisms by which neonatal E. coli transcytose across the intestinal epithelium to produce bacteremia.
Wprowadzenie
Escherichia coli is the most common cause of early-onset sepsis in newborns1,2,3. The mortality rate of neonatal E. coli bacteremia can reach 40%, and meningitis is a possible complication that is associated with severe neurodevelopmental disabilities2. The ingestion of maternal E. coli strains by the newborn can produce neonatal bacteremia; this process has been replicated in animal models2,4. Once ingested, pathogenic bacteria travel from the neonatal gut lumen across the intestinal barrier and enter the bloodstream, causing septicemia. Neonatal invasive E. coli strains that produce bacteremia vary in their ability to invade intestinal epithelial cells1,5. However, their ability to transcytose the intestinal epithelium after invasion has not been completely characterized.
This intestinal transcytosis model is a useful in vitro method to emulate bacterial passage across the intestinal epithelium. The overall goal of the methods presented in this manuscript is to compare the ability of neonatal E. coli isolates to transcytose the intestinal epithelium. The model described here utilizes T84 cells, which are immortalized human intestinal adenocarcinoma cells6,7. T84 cells are grown to confluence on a semipermeable membrane with two separate compartments. The rationale for using this technique is that, as happens in vivo, these intestinal cells polarize and develop mature tight junctions6,8. The side in contact with the membrane becomes the basal side. The opposite side of the cells becomes the apical side, resembling the intestinal lumen where ingested pathogens adhere and invade. The transwell membrane is permeable to bacteria, but the polarized intestinal cells form tight junctions, which impair bacterial paracellular movement9. Thus, this method provides the advantage of a controlled in vitro environment utilizing a human cell line to study the process of bacterial transcytosis, including the transcellular route. While other methods exist to investigate the transcytosis of bacteria across the intestinal epithelium, the transwell method presented here provides greater ease and accessibility. Alternative techniques, such as those utilizing ex vivo samples set up in Ussing chamber systems, are available. However, they utilize tissue specimens that may not be easily accessible, particularly if the research intends to study human physiology10. Intestinal organoids represent another example of an in vitro alternative for studying host-bacteria interactions11. While organoid monolayers can also be used in the transwell system to study bacterial transcytosis, they require the isolation and growth of stem cells and the use of specific growth factors to induce differentiation12. Thus, their use is more time-consuming and associated with greater costs as compared to the transwell method described in this manuscript.
The assessment of bacterial passage across the intestinal epithelium using this in vitro transwell system has been successfully performed for various pathogens. These studies have shown the utility of the transwell system using T84 cells to characterize the transcytosis of bacteria across the polarized intestinal epithelium13,14,15. However, the application of this transwell method to compare the transcytosis ability of bacteremia-producing neonatal E. coli strains has not been described in detail. This manuscript provides other researchers with a standard transwell protocol that is reliable and easy to use and does not require resources that are too expensive.
To compare the ability of neonatal invasive E. coli strains to transcytose the intestinal epithelium, the apical side of the intestinal epithelial monolayer can be infected with a known number of bacterial cells. After incubation, the medium on the basal side of the epithelium can be collected and the bacteria quantified to determine the amount of bacterial transcytosis over time. In this manuscript, the methods presented are utilized to study the transcytosis ability of neonatal E. coli clinical strains recovered from newborns hospitalized with bacteremia. The inclusion criteria for the selection of these neonatal clinical isolates for transcytosis studies have been published previously1,2,16. When this method is performed using different E. coli strains, their transcytosis abilities can be compared. Through this process, the intestinal transcytosis model provides valuable data to characterize the virulence factors of E. coli that contribute to the multistep process that culminates in the development of neonatal bacteremia.
Protokół
NOTE: Perform all the manipulations of the T84 cells, bacteria, plates, and reagents in a Biosafety Level 2 (BSL-2) safety cabinet to avoid contamination. Use separate areas and incubators for all the work involving sterile T84 cells, infected T84 cells, and E. coli. The clinical E. coli isolates tested with the methods described here were obtained following the guidelines of the Institutional Review Board at our institution1,16.
1. Preparing transcytosis inserts with T84 cells (approximately 1-2 weeks before the experiment)
- Grow American Type Culture Collection (ATCC) T84 cells in tissue culture medium (TCM + antibiotics) consisting of Dulbecco's Modified Eagle Medium:Ham's F-12 nutrient mixture (1:1, final concentration: 50% each), 5% fetal bovine serum, and 1% (100 U/mL) penicillin/streptomycin dual antibiotic mixture. Incubate the cells at 37 °C with 5% CO2.
NOTE: A variation of this medium formulation without penicillin/streptomycin (TCM w/o antibiotics) is used for the later steps (section 2 and onward) in the procedure. Ensure that the correct formulation is used for each step. - Working inside a biosafety cabinet (BSC), seed the T84 cells into polyethylene terephthalate membrane cell culture transwell inserts with 3 µm pores made for 24-well plates. Include transwell insert replicates for each desired experimental condition plus uninfected controls to monitor for possible contamination.
- In a 24-well plate designed to hold transwell inserts, fill the desired number of collecting wells with 1 mL of TCM + antibiotics.
- In each well, place one transwell insert.
- Seed these inserts with 1 x 105 T84 cells suspended in 500 µL of TCM + antibiotics. Quantify the number of cells using a hemocytometer with trypan blue stain or an automated cell counter17.
- Incubate the transwell plates containing seeded inserts in the same conditions that the cells were grown in.
- Verify with light microscopy that the monolayers have started to become confluent after seeding the insert, approximately 48 h after seeding.
- Every 2 days after seeding, measure and record the transepithelial electrical resistance (TEER) using an epithelial volt/ohm meter (EVOM) to assess the maturity of the monolayer. Once the inserts reach a TEER of at least 1,000 Ω·cm2, they are deemed ready for the assay18.
NOTE: The inserts typically take 7-10 days to reach this TEER after seeding. The T84 cells will show 100% confluence under light microscopy once this resistance is reached.- Store the EVOM probe with the electrodes submerged in 0.15 M KCl when not in use.
- Prior to measuring the TEER, decontaminate the electrode probe by submerging it in 5 mL of 70% ethanol in a 50 mL conical tube for 10-15 min. Remove the probe, shake off the excess ethanol, and allow it to air-dry inside the BSC for 10 min. Retain the tube of ethanol.
- Test the EVOM and probe by placing the dry decontaminated probe in a sterile well containing 1 mL of TCM + antibiotics with a sterile insert inside containing 500 µL of TCM + antibiotics. Ensure that the EVOM reading is <200 Ω. Record this blank value to use it in the later resistance calculations described in step 1.3.6.
- Remove the probe from the tube of TCM + antibiotics. Retain this tube for the storage of the probe throughout the experiment.
- Gently lower the probe into the first insert with the long electrode in the collecting well and the short electrode inside the insert. Allow the long electrode to touch the bottom of the collecting well, but do not push down as this may disrupt the epithelial monolayer.
- Repeat this process to measure and record the resistance in Ohms (Ω) for every insert. When finished, decontaminate the probe in the ethanol by submerging it for another 10-15 min. Then, move the decontaminated probe back to the KCl solution for storage. Subtract the blank resistance obtained in step 1.3.3 from each value obtained from each insert containing T84 cells. Multiply the resulting resistance (Ω) for each insert by the area of the bottom of each insert (cm2) to obtain the final TEER measurement (Ω·cm2).
- Once the TEER reaches at least 1,000 Ω·cm2, the epithelial monolayer is mature and ready for infection assays.
- As the TEER matures, provide the cells with fresh medium every 1-2 days.
- In a new 24-well plate, add 1 mL of TCM + antibiotics to one well for each seeded insert being prepared.
- Using sterile forceps, carefully transfer the inserts to the newly replenished wells.
- Replace the media in the inserts.
- Remove the old media from the inserts by tilting the plate and using an in-house vacuum aspirator to gently remove the media with a pipette tip along the side of the insert. The aspirator allows the regulation of low-level suction to prevent the disruption of the cells. Do not allow the pipette tip to touch the bottom of the insert, as this will disrupt the developing epithelial monolayer.
- Add 500 µL of TCM + antibiotics to the inserts. Visualize the monolayer with light microscopy to verify that it remains intact.
- Every 1-2 days, measure the TEER across each insert as described above in steps 1.3.2-1.3.6.
2. Preparing the T84 cells 1 day before the experiment using TCM w/o antibiotics
- Measure and record the TEER the day before the experiment.
- Replace the TCM in the same manner as during the prior cell preparation and maintenance. However, TCM w/o antibiotics is used instead in preparation for the infection (1 mL in the plate well and 500 µL in the insert).
3. E. coli cultures (started 1 day before the experiment)
CAUTION: Use Biosafety Level 2 (BSL-2) precautions when working with pathogenic clinical E. coli strains.
- Take a labeled 15 mL conical tube with 5 mL of sterile lysogeny broth (LB), and use a sterile loop to inoculate the broth with one colony from one bacterial strain (E. coli). Repeat this process, creating one overnight culture tube for each strain to be tested.
- Incubate the overnight culture, with the caps of the tubes loosened, in an incubator shaker (250 rpm, 37 °C).
4. Preparing the E. coli inoculum, epithelial cells, and materials (on the morning of the experiment)
NOTE: Use TCM w/o antibiotics warmed up to 37 °C from this point on.
- Add 250 µL from each overnight LB culture to 25 mL of TCM w/o antibiotics in a 50 mL conical tube (one per individual strain). Keep the caps of the tubes loosened. Place these new culture tubes in the shaker at the same settings (250 rpm, 37 °C) for exactly 2 h. Perform the remaining substeps while waiting.
- Measure the TEER across each insert, as described in steps 1.3.2-1.3.6. Record these as the TEERs at time (t) = 0 h.
- Move the inserts to wells in a new plate, and change the media using the technique described in step 1.4.3. However, this time, fill the new collecting wells with 500 µL of TCM w/o antibiotics, and fill the inserts with 400 µL of TCM w/o antibiotics. Keep the inserts inside the tissue culture incubator until the time of infection.
- Set out a sufficient number of square LB agar plates to warm to room temperature (RT) for later plating and bacterial quantification.
5. Inoculation of the cells (start of the experiment)
- After exactly 2 h, remove the morning bacterial cultures from the shaker, and centrifuge them for 10 min (1,900 x g, 4 °C).
NOTE: For all the following steps, keep all the bacterial suspensions on ice to minimize growth. - Resuspend the bacteria pellet in TCM w/o antibiotics. Use a spectrophotometer to adjust the optical density (OD) to 0.7-0.9, and further dilute with TCM w/o antibiotics to a concentration of 1 x 106 colony forming units (CFU)/mL (approximately 1:100 dilution). Use this bacterial suspension to infect each insert with 1 x 105 CFU per 100 µL volume.
- Label the transwell plate, and infect each insert with 100 µL of the OD-adjusted inoculum (total of 1 x 105 CFU per insert). The assay has now begun. Note the time, and record it as t = 0 h.
- Plate the inoculum bacterial suspension to quantify the CFU/mL using the track dilution method, plating 10 µL aliquots on square LB agar plates19.
6. Quantifying transcytosis
- Every 30 min following inoculation, fill new wells with 500 µL of TCM w/o antibiotics. Transfer the inserts to these new wells using a different set of sterile forceps for each different bacterial strain.
- Collect the media from the used collecting well for each insert into separate labeled tubes. Place these tubes on ice. Return the transwell plates to the incubator in between time points.
- For each insert, combine the collected media from t = 0.5 h, t = 1 h, t = 1.5 h, and t = 2 h, and vortex briefly. Plate the collected media on LB agar plates using the track dilution method to quantify the amount of bacteria transcytosed in the first 2 h of the experiment.
NOTE: Retrieving the bacteria every 30 min and keeping them on ice ensures the bacterial growth in the collecting wells is minimized and measurements are done on predominantly transcytosed bacteria. - Place the labeled track dilution LB agar plates in the bacterial incubator at 37 °C without supplemental CO2, and return the T84 transwell plate to the tissue culture incubator.
- At t = 4 h, repeat step 6.3 by combining the collected media from t = 2.5 h, t = 3 h, t = 3.5 h, and t = 4 h.
- At t = 6 h, repeat step 6.3 by combining the collected media from t= 4.5 h, t = 5 h, t = 5.5 h, and t = 6 h. Additionally, at t = 6 h, plate the media from the control wells.
7. End of the experiment
- Measure and record the TEER at the end of the experiment, t = 6 h. Use the procedure described in steps 1.3.2-1.3.6.
- Decontaminate the probe by submerging it in 70% ethanol for 10-15 min. Discard the inserts, or save/process them for additional applications, if desired. Allow the LB agar plates to incubate overnight, and disinfect and/or safely dispose of all the other used materials.
- After overnight incubation, count the bacterial colonies manually on the track dilution LB plates to determine the inoculum amount and the amount of E. coli transcytosis. Ensure that the control plates do not show any bacterial growth.
Wyniki
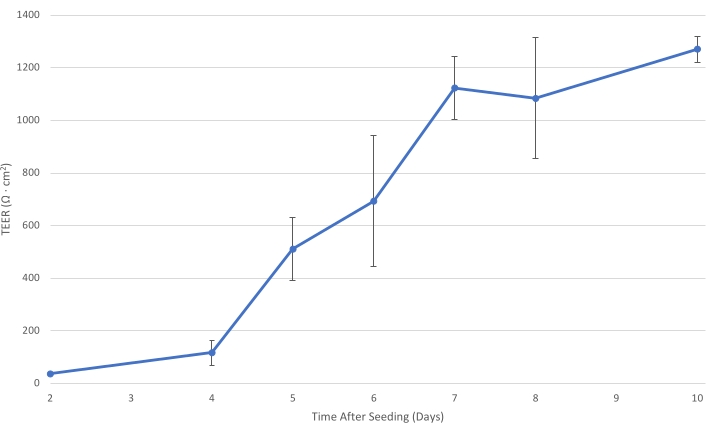
Figure 1: T84 TEER over time. As the T84 cell layer matures on the insert, the electrical resistance of the monolayer increases. At a TEER of at least 1,000 Ω·cm2, the cell layer is sufficiently developed to decrease the paracellular bacterial transport and allow the measurement of primarily transcellular bacterial transit....
Dyskusje
This method is derived from techniques used in gastroenterology and infectious disease20. In vitro models of the intestinal epithelial barrier have been used to elucidate the mechanisms by which the luminal contents interact with this relevant component of innate immunity6,8. The host-pathogen interactions of invasive neonatal E. coli have also been separately characterized through genetic analysis, studies of antimicrobi...
Ujawnienia
None.
Podziękowania
This work was supported by a Sarah Morrison student grant issued by the University of Missouri-Kansas City School of Medicine to A.I.
Materiały
| Name | Company | Catalog Number | Comments |
| 10,000 U/ mL Penicillin/Streptomycin Mixture | Fisher Scientific | 15-140-122 | |
| 15 mL sterile conical tubes | MidSci | C15B | |
| 2 mL microcentrifuge tubes | Avant | AVSS2000 | |
| 50 mL sterile polypropylene conical tubes | Falcon | 352070 | |
| Aspirator | Corning | 4930 | |
| Biosafety Cabinets | Labconco | 30441010028343 | Three of these are used in the method: one for sterile tissue work, one for infected tissue work, and one for bacterial work. |
| Centrifuge | Sorvall | Legend RT | |
| Disposable inoculation loops | Fisherbrand | 22363605 | |
| Dulbecco's Modified Eagle Medium (DMEM) | Gibco | 11965-084 | |
| Epithelial Volt/Ohm Meter | World Precision Instruments | EVOM2 | |
| Fetal Bovine Serum | Fisher Scientific | 10437028 | |
| Ham's F-12 Nutrient Mixture | Gibco | 11765-047 | |
| Hemacytometer | Sigma Aldrich, Bright Line | Z359629 | |
| Incubator shaker | New Brunswick | Innova 4080 | |
| Incubators | Thermo Scientific | 51030284 | Three of these are used in the method: one for sterile tissue culturing, one for infected tissue culturing, and one for bacterial incubation. |
| Lysogeny broth | Difco | 244610 | |
| Lysogeny broth agar | IBI Scientific | IB49101 | |
| Nikon Eclipse TS2R Microscope | Nikon | ||
| Spectrophotometer | Unico | 1100RS | |
| T84 Intestinal Cells | American Tissue Culture Collection | CCL248 | |
| Tissue culture inserts, with polyethylene trephthalate membrane, 3 µm pores, 24 well format | Falcon | 353096 | |
| Tissue culture plate, 24 wells | Falcon | 353504 | |
| Trypan blue stain | Fisher Scientific | T10282 |
Odniesienia
- Shakir, S. M., Goldbeck, J. M., Robison, D., Eckerd, A. M., Chavez-Bueno, S. Genotypic and phenotypic characterization of invasive neonatal Escherichia coli clinical isolates. American Journal of Perinatology. 31 (11), 975-982 (2014).
- Cole, B. K., et al. Route of infection alters virulence of neonatal septicemia Escherichia coli clinical isolates. PloS One. 12 (12), 0189032 (2017).
- Stoll, B. J., et al. Early-onset neonatal sepsis 2015 to 2017, the rise of Escherichia coli, and the need for novel prevention strategies. Journal of the American Medical Association Pediatrics. 174 (7), 200593 (2020).
- Dalgakiran, F., Witcomb, L. A., McCarthy, A. J., Birchenough, G. M., Taylor, P. W. Non-invasive model of neuropathogenic Escherichia coli infection in the neonatal rat. Journal of Visualized Experiments. (92), e52018 (2014).
- Williams, M., et al. Whole-genome sequencing-based phylogeny, antibiotic resistance, and invasive phenotype of Escherichia coli strains colonizing the cervix of women in preterm labor. BMC Microbiology. 21 (1), 330 (2021).
- Schoultz, I., Keita, &. #. 1. 9. 7. ;. The intestinal barrier and current techniques for the assessment of gut permeability. Cells. 9 (8), 1909 (2020).
- Devriese, S., et al. T84 monolayers are superior to Caco-2 as a model system of colonocytes. Histochemistry and Cell Biology. 148 (1), 85-93 (2017).
- Buckley, A., Turner, J. R. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harbor Perspectives in Biology. 10 (1), 029314 (2018).
- Awad, W. A., Hess, C., Hess, M. Enteric pathogens and their toxin-induced disruption of the intestinal barrier through alteration of tight junctions in chickens. Toxins. 9 (2), 60 (2017).
- Vancamelbeke, M., Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert Review of Gastroenterology & Hepatology. 11 (9), 821-834 (2017).
- Aguilar, C., et al. Organoids as host models for infection biology - A review of methods. Experimental and Molecular Medicine. 53 (10), 1471-1482 (2021).
- Nickerson, K. P., et al. A versatile human intestinal organoid-derived epithelial monolayer model for the study of enteric pathogens. Microbiology Spectrum. 9 (1), 0000321 (2021).
- Gavin, H. E., Beubier, N. T., Satchell, K. J. The effector domain region of the Vibrio vulnificus MARTX toxin confers biphasic epithelial barrier disruption and is essential for systemic spread from the intestine. PLoS Pathogens. 13 (1), 1006119 (2017).
- Kobayashi, H., et al. Aeromonas sobria serine protease decreases epithelial barrier function in T84 cells and accelerates bacterial translocation across the T84 monolayer in vitro. PloS One. 14 (8), 0221344 (2019).
- Kalischuk, L. D., Inglis, G. D., Buret, A. G. Campylobacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut Pathogens. 1 (1), 2 (2009).
- Cole, B. K., Ilikj, M., McCloskey, C. B., Chavez-Bueno, S. Antibiotic resistance and molecular characterization of bacteremia Escherichia coli isolates from newborns in the United States. PloS One. 14 (7), 0219352 (2019).
- Cadena-Herrera, D., et al. Validation of three viable-cell counting methods: Manual, semi-automated, and automated. Biotechnology Reports. 7, 9-16 (2015).
- den Hartog, G., et al. Apurinic/apyrimidinic endonuclease 1 restricts the internalization of bacteria into human intestinal epithelial cells through the inhibition of Rac1. Frontiers in Immunology. 11, 553994 (2020).
- Jett, B. D., Hatter, K. L., Huycke, M. M., Gilmore, M. S. Simplified agar plate method for quantifying viable bacteria. Biotechniques. 23 (4), 648-650 (1997).
- Lievin-Le Moal, V., Servin, A. L. Pathogenesis of human enterovirulent bacteria: Lessons from cultured, fully differentiated human colon cancer cell lines. Microbiology and Molecular Biology Reviews. 77 (3), 380-439 (2013).
- Kaczmarek, A., Budzynska, A., Gospodarek, E. Detection of K1 antigen of Escherichia coli rods isolated from pregnant women and neonates. Folia Microbiologica. 59 (5), 419-422 (2014).
- Kalita, A., Hu, J., Torres, A. G. Recent advances in adherence and invasion of pathogenic Escherichia coli. Current Opinion in Infectious Diseases. 27 (5), 459-464 (2014).
- McCool, D. J., Marcon, M. A., Forstner, J. F., Forstner, G. G. The T84 human colonic adenocarcinoma cell line produces mucin in culture and releases it in response to various secretagogues. Biochemical Journal. 267 (2), 491-500 (1990).
- Resta-Lenert, S., Barrett, K. E. Enteroinvasive bacteria alter barrier and transport properties of human intestinal epithelium: Role of iNOS and COX-2. Gastroenterology. 122 (4), 1070-1087 (2002).
- Elatrech, I., et al. Escherichia coli LF82 differentially regulates ROS production and mucin expression in intestinal epithelial T84 cells: Implication of NOX1. Inflammatory Bowel Diseases. 21 (5), 1018-1026 (2015).
- El-Aouar Filho, R. A., et al. Heterogeneous family of cyclomodulins: Smart weapons that allow bacteria to hijack the eukaryotic cell cycle and promote infections. Frontiers in Cellular and Infection Microbiology. 7, 208 (2017).
- Hopkins, A. M., Walsh, S. V., Verkade, P., Boquet, P., Nusrat, A. Constitutive activation of Rho proteins by CNF-1 influences tight junction structure and epithelial barrier function. Journal of Cell Science. 116, 725-742 (2003).
- Shiou, S. R., et al. Erythropoietin protects intestinal epithelial barrier function and lowers the incidence of experimental neonatal necrotizing enterocolitis. Journal of Biological Chemistry. 286 (14), 12123-12132 (2011).
- Newburg, D. S., Ko, J. S., Leone, S., Nanthakumar, N. N. Human milk oligosaccharides and synthetic galactosyloligosaccharides contain 3’-, 4-, and 6'-galactosyllactose and attenuate inflammation in human T84, NCM-460, and H4 cells and intestinal tissue ex vivo. Journal of Nutrition. 146 (2), 358-367 (2016).
- Burns, J. L., Griffith, A., Barry, J. J., Jonas, M., Chi, E. Y. Transcytosis of gastrointestinal epithelial cells by Escherichia coli K1. Pediatric Research. 49 (1), 30-37 (2001).
- Raut, B., Chen, L. J., Hori, T., Kaji, H. An open-source add-on EVOM((R)) device for real-time transepithelial/endothelial electrical resistance measurements in multiple transwell samples. Micromachines. 12 (3), 282 (2021).
- McCarthy, A. J., Stabler, R. A., Taylor, P. W. Genome-wide identification by transposon insertion sequencing of Escherichia coli K1 genes essential for in vitro growth, gastrointestinal colonizing capacity, and survival in serum. Journal of Bacteriology. 200 (7), 00698 (2018).
- Sayoc-Becerra, A., et al. The JAK-inhibitor tofacitinib rescues human intestinal epithelial cells and colonoids from cytokine-induced barrier dysfunction. Inflammatory Bowel Diseases. 26 (3), 407-422 (2020).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone