A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Induction of Murine Intestinal Inflammation by Adoptive Transfer of Effector CD4+CD45RBhigh T Cells into Immunodeficient Mice
In This Article
Summary
Here, we present a protocol to induce colonic inflammation in mice by adoptive transfer of syngeneic CD4+CD45RBhigh T cells into T and B cell deficient recipients. Clinical and histopathological features mimic human inflammatory bowel diseases. This method allows the study of the initiation of colonic inflammation and progression of disease.
Abstract
There are many different animal models available for studying the pathogenesis of human inflammatory bowel diseases (IBD), each with its own advantages and disadvantages. We describe here an experimental colitis model that is initiated by adoptive transfer of syngeneic splenic CD4+CD45RBhigh T cells into T and B cell deficient recipient mice. The CD4+CD45RBhigh T cell population that largely consists of naïve effector cells is capable of inducing chronic intestinal inflammation, closely resembling key aspects of human IBD. This method can be manipulated to study aspects of disease onset and progression. Additionally it can be used to study the function of innate, adaptive, and regulatory immune cell populations, and the role of environmental exposures, i.e., the microbiota, in intestinal inflammation. In this article we illustrate the methodology for inducing colitis with a step-by-step protocol. This includes a video demonstration of key technical aspects required to successfully develop this murine model of experimental colitis for research purposes.
Introduction
The inflammatory bowel diseases (IBD) Crohn’s disease and ulcerative colitis result from an incompletely defined and complex interaction between host immune responses, genetic susceptibility, environmental factors, and the enteric luminal contents1. Recent genome-wide association studies report associations between immune cell regulatory genes and IBD susceptibility2,3. Both innate and adaptive immune cell intrinsic genes are represented in these studies, indicating a central role for these cell populations in IBD pathogenesis.
There currently exist more than 50 animal models of human IBD. While no one model perfectly phenocopies human IBD, many are useful for studying various aspects of human disease, including disease onset and progression and the wound-healing response. In the method described here, intestinal inflammation is initiated with syngeneic splenic CD4+CD45RBhigh T cell adoptive transfer into T and B cell deficient recipient mice4. The CD4+CD45RBhigh T cell population contains mainly naïve T cells primed for activation that are capable of inducing chronic small bowel and colonic inflammation. This method allows the researcher to modify key experimental variables, including both innate and adaptive immune cell populations, to answer biologically relevant questions relating to disease pathogenesis. Additionally, this method provides precise initiation of disease onset and a well-characterized experimental time course. This permits the kinetic study of clinical features of disease progression in mice. Intestinal inflammation induced by this method shares many features with human IBD, including chronic large and small bowel transmural inflammation, pathogenesis driven by cytokines such as TNF and IL-12, and systemic symptoms such as wasting5. Thus, it is an ideal model system for studying the pathogenesis of human IBD.
The method here describes in detail the protocol for inducing experimental colitis by adoptive transfer of CD4+CD45RBhigh T cells into Rag1-/- mice. We discuss key technical steps, expected results, optimization, and trouble-shooting. We address the required elements for the successful development of this murine model of intestinal inflammation for research purposes.
Access restricted. Please log in or start a trial to view this content.
Protocol
NOTE: Ensure that all animal protocols are approved by and in compliance with Institutional Animal Care and Use Committee (IACUC) regulations and the National Research Council’s Guide for the Care and Use of Laboratory Animals. Donor mice may be either male or female, but recipient mice should be male. If female recipients are to be used, the donor mice must be female5. Maintain colonies using regular, non-sterile bedding and non-acidified water, as these may impact the intestinal microbiota, and, thus, the colitis phenotype of the mice5,6.
1. Experimental Preparation
- Use ice-cold media and buffers. Keep cells on ice throughout the experiment.
- Perform the experiment in sterile biohazard hood using sterile technique.
2. Isolation of Splenic T cells
- Euthanize donor mouse/mice in CO2 chamber followed by cervical dislocation. Spray abdomen with 70% ethanol.
- Make a horizontal incision in the abdomen and peel away skin to expose peritoneum. Hold peritoneum away from the internal organs with the forceps and make an incision in the left abdominal peritoneum to expose and excise the spleen.
- Place the spleen in 10 ml of Complete Media in a Petri dish. Remove and discard excess tissue from spleen.
- Use 2 sterilized glass slides to crush and tease apart spleen into single-cell suspension. Filter cell suspension through a 70 μm strainer into a 50 ml conical tube and rinse strainer with 5 ml of Complete Media. Place up to 5 spleens in one 50 ml conical tube.
- Centrifuge cells at 450 x g for 7 min. Discard supernatant by pouring off or by vacuum suction into waste container.
- Gently resuspend cells in 5 ml per spleen of Lysis Buffer to lyse red blood cells for 10 min at room temperature. Add an equal volume of Complete Media (5 ml per spleen) to the tube.
- Centrifuge cells at 450 x g for 7 min. Discard supernatant by pouring off or by vacuum suction into waste container.
- Gently resuspend cells in 10 ml of Labeling Buffer.
- Count cells by trypan blue exclusion.
- Remove 20 μl of cell suspension and add to 180 μl trypan blue to a microfuge tube and mix thoroughly. After 5 min, add 10 μl labeled cells to hemocytometer and count non-blue cells under the microscope. Determine total number of viable cells. Discard cell/trypan blue mix.
- Centrifuge cells in 10 ml of Labeling Buffer at 450 x g for 7 min. Discard supernatant by pouring off or by vacuum suction into waste container.
3. Enrichment of CD4+ T cells
NOTE: Follow manufacturer’s instructions for specific products used in this section.
- Gently resuspend cells to a single-cell suspension of 20 x 106 cell/ml in cold Labeling Buffer.
- Add 5 μl per 1 x 106 cells of biotinylated CD4 T cell enrichment antibodies. Incubate cells on ice for 15 min.
- Add 10x volume of Labeling Buffer. Centrifuge cells at 450 × g for 7 min. Carefully aspirate all the supernatant using vacuum suction into waste container.
- Thoroughly vortex streptavidin-conjugated magnetic particles. Add 5 μl of particles per 1 x 106 cells.
- Mix thoroughly but gently. Keep mix at 6-12 °C for 30 min.
- Add Labeling Buffer to a concentration of 20-80 x 106 cells/ml. Transfer up to 1.0 ml labeled cells per 12 x 75 mm round-bottom test tube (referred to as the “positive-fraction tube”).
- Place each positive-fraction tube on magnet for 6-8 min.
- With the positive-fraction tubes still on the magnet, carefully transfer supernatant from positive-fraction tube with glass Pasteur pipet to a new sterile 50 ml conical tube (referred to as the “enriched fraction”). This enriched fraction contains CD4+ T cells. Be careful not to disrupt the labeled cells attracted to the magnet.
- Resuspend cells left in the positive-fraction tubes in the same volume of Labeling Buffer as in step 3.6 by pipetting up and down vigorously. Place positive-fraction tube back on magnet for 6-8 min.
- With positive-fraction tubes still on the magnet, carefully transfer supernatant (enriched fraction, CD4+) from positive-fraction tube with glass Pasteur pipet to sterile 50 ml conical tube from step 3.8 without disrupting cells attached to magnet.
- Repeat steps 3.9-3.10 to increase the yield of CD4+ T cells obtained. Continue protocol using enriched fraction (CD4+ cells).
4. Labeling and Sorting Cells7
- Centrifuge enriched cells at 450 x g for 7 min. Discard supernatant by pouring off or vacuum suction into waste container.
- Resuspend cells in 1 ml of Labeling Buffer. Remove aliquot of cells to count and to assess for cell viability by trypan blue exclusion as in step 2.9.
- Add a volume of Labeling Buffer to 10 x 106 cells/ml; if cells are already <10 x 106, centrifuge at 450 x g for 7 min, discard supernatant by pouring off or vacuum suction into waste container, and add volume of Labeling Buffer to 10 x 106 cells/ml.
- Set up separate aliquots of approximately 5-10 × 105 cells each for unstained, isotype-stained, and single-stained control cells in microfuge tubes.
- Add 5 μg/ml CD4-FITC and 1 μg/ml CD45RB-PE to cells. Add isotype control stains and single stains at the same concentration to appropriate aliquots in microfuge tubes. Mix well but gently and incubate on ice protected from light for 30 min.
- Add 10x volume of Labeling Buffer to cells and controls. Centrifuge 450 x g for 7 min. Discard supernatant by vacuum suction into waste container.
- Resuspend in Labeling Buffer to volume in step 4.5. Centrifuge 450 x g for 7 min. Discard supernatant by vacuum suction into waste container.
- Resuspend in Labeling Buffer to 10 x 106 cells/ml. Pass cells through 70 μm strainer into FACS tube. Keep on ice protected from light until ready for FACS.
- Set up and determine appropriate compensation on the cell sorter with unstained cells and single-stained controls.
- Exclude nonviable cells with forward- and side-scatter gating (Figure 1A). Set up gating for CD4+ and CD45RB+ cells with isotype-stained controls. Gate cells on CD4+ population.
- Sort CD4+ cells into CD45RBhigh and CD45RBlow populations using a simple histogram for PE-stained cells into tubes with 2 ml of Complete Media. The CD45RBhigh population represents the highest 40% of CD4+CD45RB+ cells (CD45RBhigh), and the CD45RBlow population is the lowest 20% of CD4+CD45RB+ cells (CD45RBlow; Figure 1B).
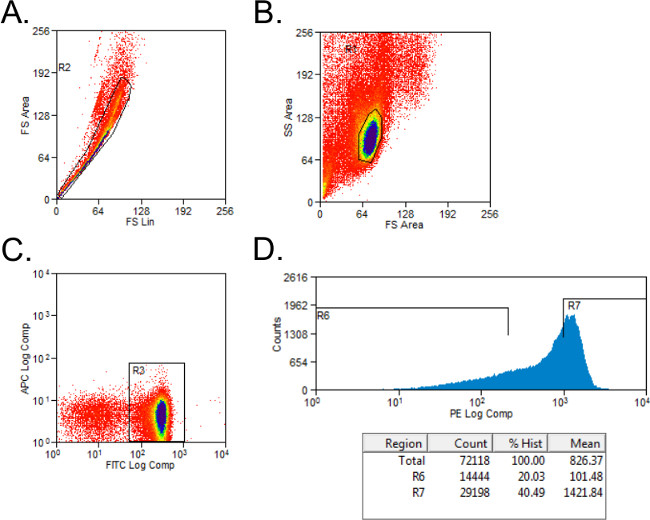
Figure 1: Representative flow cytometry plots of CD4+CD45RB T cell populations during FACS analysis. (A-C) FITC CD4- and PE CD45RB-stained splenocytes from donor C57BL/6 mice were sorted by FACS into CD4+CD45RBhigh and CD4+CD45RBlow T cell populations. (A) Doublet events were excluded on the forward scatter plot. (B) Lymphocytes were gated in the forward and side scatter plot. (C) CD4+ T cells were gated, and (D) CD4+CD45RB+ T cells were plotted on a PE versus Events histogram. CD4+CD45RBlow cells were considered to be the lowest 20% of CD45RB+ cells. CD4+CD45RBhigh cells were defined as the highest 40% of CD45RB+ cells.
- Run an aliquot of each cell population on the FACS machine to assess purity of populations.
- Centrifuge sorted cells at 450 x g for 7 min. Resuspend in 1 ml PBS. Remove aliquot of cells to count and to assess for cell viability by trypan blue exclusion as in step 2.9.
5. Injection of Cells into Recipients
- Resuspend sorted cells to 4 x 106 cells/ml (CD45RBhigh) or 2 x 106 cells/ml (CD45RBlow) in PBS.
- Transfer 100 μl of CD45RBhigh cells per recipient to new sterile tube. Add 100 μl of PBS per recipient to this tube. Thus, total injection volume per animal is 200 μl, and total amount of cells per recipient is 4 x 105 CD4+CD45RBhigh naïve T cells.
- If experimental group receiving T regulatory cells is desired, transfer 100 μl of CD45RBhigh cells per recipient to new sterile tube. Add 100 μl of CD45RBlow cells per recipient to the same tube. Total injection volume per animal is 200 μl; ratio of CD45RBhigh:CD45RBlow cells is 2:1.
- Inject 100 μl of CD45RBhigh or CD45RBhigh/CD45RBlow CD4+ cells intraperitoneally into the right and left side of the abdomen (total of 200 μl) of each recipient.
6. Monitoring of Disease Progression in Recipient Animals
- To assess the clinical status of the recipient animals, assign aggregate clinical scores for the following parameters8 on the day of injection, weekly thereafter, and at time of sacrifice:
- Determine wasting by measuring weight loss: 0 – no weight loss; 1 – 0.1-10% loss of initial body weight; 2 – more than 10% loss of initial body weight (Figure 2A).

Figure 2: Clinical and gross pathological signs of inflammation occur after transfer of wild type CD4+CD45RBhigh T cells into Rag1-/- and NRDKO recipient mice11. (A) NRDKO recipients lost on average 10% of their initial body weights by 5 weeks post-transfer, whereas Rag1-/- recipients did not exhibit clinical signs of disease at this time. Each point represents the mean percentage of initial body weight for the cohort ± SEM. **, p <0.005. (B) Some mice developed severe intestinal inflammation, as demonstrated by the presence of rectal prolapse. This is a representative picture of rectal prolapse in a NRDKO recipient mouse. (C) Grossly, colons from both Rag1-/- and NRDKO recipient mice are thickened and shortened compared to colons from Rag1-/- and NRDKO mice without T cell adoptive transfer. Colons from NRDKO recipient mice show severe inflammation and increased colon weights (data not shown). Please click here to view a larger version of this figure.
-
- Determine quality of stool by placing animal in clean container until it has passed stool: 0 – none; 1 – soft stool; 2 – watery and/or bloody stool.
- Determine general health of animal by presence of the following signs of disease: 0 – no hunched posture, bristled fur, or skin lesions; 1 – any one of the following present: hunched posture, bristled fur, or skin lesions.
- Determine presence of rectal prolapse: 0 – absent; 1 – present (Figure 2B).
- Sacrifice animals by CO2 inhalation followed by cervical dislocation when they have lost 20% of their starting body weight or at desired time point, whichever comes first. Clinical disease is typically apparent starting at week 5 post-repletion.
- Assess for disease severity as previously described5,7.
- Assign clinical scores as in step 6.18.
- Measure colon length and weight (Figure 2C).
- Perform histological analysis of inflammation5.
- Determine spontaneous cytokine expression in intestinal tissue explant cultures9, mesenteric lymph nodes10, and/or serum11.
- Briefly, for explant cultures9, remove colons after sacrifice, open longitudinally, and clean with PBS. Incubate colons on an orbital shaker set at 250 rpm in Complete Media for 30 min at room temperature.
- Chop tissue into small pieces and incubated at 37 °C in Complete Media for 24 hr. Collect the supernatant and use for quantitation of cytokines per 100 mg tissue by ELISA.
- Perform ex vivo characterization of T cell phenotypes and/or function10.
Access restricted. Please log in or start a trial to view this content.
Results
Approximately 10 x 106 CD4+CD45RBhigh T cells from 10 spleens from adult C57BL/6 donor mice are reliably isolated. This number will vary depending on the age and strain of the donor mouse and the proficiency of the researcher. When 4 x 105 C57BL/6 CD4+CD45RBhigh T cells are transferred into C57BL/6 Rag1-/- recipient mice, clinical signs of disease emerge around week 5 post-repletion or sooner if mice are genetically susceptible to more ...
Access restricted. Please log in or start a trial to view this content.
Discussion
Here we describe a step-by-step protocol inducing colonic inflammation in mice by adoptive transfer of CD4+CD45RB+ T cells into immunodeficient mice. We used C57BL/6 donor spleens and syngeneic Rag1-/- recipient mice, although other strains (e.g., BALB/c, 129S6/SvEv, non-obese diabetic (NOD)) and genetic models of immunodeficiency (e.g., SCID, Rag2-/-) may also be used4,14-16. It is well established that background strain affects e...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgements
This work was supported by American Gastroenterological Association (AGA) Research Scholars Award and Crohn’s and Colitis Foundation of America (CCFA) Career Development Award (to S.Z.S.), NIH NIDDK F30 DK089692 (to E.C.S.), and University of North Carolina Center for Gastrointestinal Biology and Disease Grant P30 DK34987 (Histology Core). The UNC Flow Cytometry Core Facility is supported in part by an NCI Center Core Support Grant (P30CA016086) to the UNC Lineberger Comprehensive Cancer Center. We thank Luke B. Borst from North Carolina State University College of Veterinary Medicine for his help with histopathological analysis and immunohistochemistry.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Name of Reagent/ Equipment | Company | Catalog Number | Comments/Description |
| 10x PBS | Gibco | 14200075 | |
| 12 mm x 75 mm round-bottom tube | Falcon | 352052 | |
| 15 ml conical | Corning | 430790 | |
| 26 G x 3/8 Needle | BD Biosciences | 305110 | |
| 50 ml conical | Corning | 430828 | |
| 70 μm Cell Strainer | Fisherbrand | 22363548 | |
| BD IMagnet | BD Biosciences | 552311 | |
| β-mercaptoethanol | Thermo Scientific | 35602 | |
| CD4-FITC IgG2b | eBioscience | 11-0041 | |
| CD45RB-PE IgG2a | BD Pharminogen | 553101 | |
| Complete Media | RPMI-1640, 1% Pen/Strep, 10% FBS, 0.0004% β-ME | ||
| FACS tube + strainer | BD Falcon | 352235 | |
| Glass Microscope Slides | Fisherbrand | 12550A3 | |
| Heat-inactivated FBS | Gemini | 100-106 | |
| Labeling Buffer | 1x PBS, 0.5% BSA, 2 mM EDTA | ||
| Lysis Buffer | 0.08% NH4Cl, 0.1% KHCO3, 1 mM EDTA | ||
| MoFlo XDP | Beckman Coulter | ||
| Mouse CD4 T lymphocyte Enrichment Set - DM | BD Biosciences | 558131 | |
| Mouse IgG2a-PE | BD Pharminogen | 553457 | |
| Mouse IgG2b-FITC | eBioscience | 11-4732 | |
| Pasteur pipet | Fisherbrand | 13-678-20D | |
| Penicillin-Streptomycin Solution, 100X | Corning Cellgro | 30-002-CI | |
| [header] | |||
| Petri Dish | Fisherbrand | 875713 | |
| Pure Ethanol 200 Proof | Decon Labs | 2705-HC | |
| RPMI-1640 | Gibco | 11-875-093 | |
| Syringe | BD Biosciences | 309597 | |
| Trypan blue | Corning Cellgro | 25-900-CI | |
| Wash Media | RPMI-1640, 1% Pen/Strep, 0.0004% β-ME | ||
References
- Xavier, R. J., Podolsky, D. K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 448 (7152), 427-434 (2007).
- Cho, J. H., Brant, S. R. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 140 (6), 1704-1712 (2011).
- Jostins, L., et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 491 (7422), 119-124 (2012).
- Powrie, F., Leach, M. W., Mauze, S., Caddle, L. B., Coffman, R. L. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol. 5 (11), 1461-1471 (1993).
- Ostanin, D. V., et al. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol. 296 (2), 135-146 (2009).
- Ma, B. W., et al. Routine habitat change: a source of unrecognized transient alteration of intestinal microbiota in laboratory mice. PLoS One. 7 (10), e47416(2012).
- Read, S., Powrie, F. Induction of inflammatory bowel disease in immunodeficient mice by depletion of regulatory T cells. Curr Protoc Immunol. Chapter 15 (Unit 15 13), (1999).
- Maillard, M. H., et al. The Wiskott-Aldrich syndrome protein is required for the function of CD4(+)CD25(+)Foxp3(+) regulatory T cells. J Exp Med. 204 (2), 381-391 (2007).
- Hegazi, R. A., et al. Carbon monoxide ameliorates chronic murine colitis through a heme oxygenase 1-dependent pathway. J Exp Med. 202 (12), 1703-1713 (2005).
- Kole, A., et al. Type I IFNs regulate effector and regulatory T cell accumulation and anti-inflammatory cytokine production during T cell-mediated colitis. J Immunol. 191 (5), 2771-2779 (2013).
- Kobayashi, T., et al. NFIL3-deficient mice develop microbiota-dependent, IL-12/23-driven spontaneous colitis. J Immunol. 192 (4), 1918-1927 (2014).
- Steinbach, E. C., et al. Innate PI3K p110delta Regulates Th1/Th17 Development and Microbiota-Dependent Colitis. J Immunol. 192 (8), 3958-3968 (2014).
- Kobayashi, T., et al. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J Immunol. 186 (8), 4649-4655 (2011).
- Leach, M. W., Bean, A. G., Mauze, S., Coffman, R. L., Powrie, F. Inflammatory bowel disease in C.B-17 scid mice reconstituted with the CD45RBhigh subset of CD4+ T cells. Am J Pathol. 148 (5), 1503-1515 (1996).
- Powrie, F., et al. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4. T cells. Immunity. 1 (7), 553-562 (1994).
- Read, S., Malmstrom, V., Powrie, F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. 192 (2), 295-302 (2000).
- Rogers, G. B., et al. Functional divergence in gastrointestinal microbiota in physically-separated genetically identical mice. Sci Rep. 4, 5437(2014).
- Fukata, M., et al. The myeloid differentiation factor 88 (MyD88) is required for CD4+ T cell effector function in a murine model of inflammatory bowel disease. J Immunol. 180 (3), 1886-1894 (2008).
- Kurtz, C. C., et al. Extracellular adenosine regulates colitis through effects on lymphoid and nonlymphoid cells. Am J Physiol Gastrointest Liver Physiol. 307 (3), 338-346 (2014).
- Naganuma, M., et al. Cutting edge: Critical role for A2A adenosine receptors in the T cell-mediated regulation of colitis. J Immunol. 177 (5), 2765-2769 (2006).
- Ranatunga, D. C., et al. A protective role for human IL-10-expressing CD4+ T cells in colitis. J Immunol. 189 (3), 1243-1252 (2012).
- Srikrishna, G., et al. Carboxylated glycans mediate colitis through activation of NF-kappa. B. J Immunol. 175 (8), 5412-5422 (2005).
- Wang, F., et al. IFN-gamma-induced TNFR2 expression is required for TNF-dependent intestinal epithelial barrier dysfunction. Gastroenterology. 131 (4), 1153-1163 (2006).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved