A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Producing Gene Deletions in Escherichia coli by P1 Transduction with Excisable Antibiotic Resistance Cassettes
In This Article
Summary
Here we present a protocol for the use of pre-existing antibiotic resistance-cassette deletion constructs as a basis for making deletion mutants in other E. coli strains. Such deletion mutations can be mobilized and inserted into the corresponding locus of a recipient strain using P1 bacteriophage transduction.
Abstract
A first approach to study the function of an unknown gene in bacteria is to create a knock-out of this gene. Here, we describe a robust and fast protocol for transferring gene deletion mutations from one Escherichia coli strain to another by using generalized transduction with the bacteriophage P1. This method requires that the mutation be selectable (e.g., based on gene disruptions using antibiotic cassette insertions). Such antibiotic cassettes can be mobilized from a donor strain and introduced into a recipient strain of interest to quickly and easily generate a gene deletion mutant. The antibiotic cassette can be designed to include flippase recognition sites that allow the excision of the cassette by a site-specific recombinase to produce a clean knock-out with only a ~100-base-pair-long scar sequence in the genome. We demonstrate the protocol by knocking out the tamA gene encoding an assembly factor involved in autotransporter biogenesis and test the effect of this knock-out on the biogenesis and function of two trimeric autotransporter adhesins. Though gene deletion by P1 transduction has its limitations, the ease and speed of its implementation make it an attractive alternative to other methods of gene deletion.
Introduction
A common first approach to study the function of a gene is to perform knock-out mutagenesis and observe the resulting phenotype. This is also termed reverse genetics. The bacterium E. coli has been the workhorse of molecular biology for the last 70 years or so, due to the ease of its culturing and its amenability to genetic manipulation1. Several methods have been developed to produce gene deletions in E. coli, including marker exchange mutagenesis2,3 and, more recently, recombineering using the λ Red or Rac ET systems4,5,6.
In a widely used system, coding sequences of individual genes are replaced by an antibiotic resistance cassette that can later be excised from the chromosome5,7. The coding sequences are replaced, for instance by a kanamycin (Kan) resistance cassette, which is flanked by flippase (FLP) recognition target (FRT) sites on either side. The FRT sites are recognized by the recombinase FLP, which mediates site-specific recombination between the FRT sites leading to the deletion of the Kan cassette. In this way, a full deletion of a given gene's coding sequence can be achieved, leaving behind only a minimal scar sequence of approximately 100 base pairs (bp) (Figure 1).
Just over a decade ago, the so-called Keio collection was developed. This is a bacterial library based on a standard laboratory E. coli K12 strain, where almost all non-essential genes were individually deleted by λ Red recombination7,8. The clones within this collection each have one coding sequence replaced with an excisable Kan resistance cassette. The Keio collection has proven to be a useful tool for many applications9. One such application is the production of deletion mutants in other E. coli strains. The Kan cassette from a given deletion clone can be mobilized by generally transducing bacteriophages, such as P110,11,12,13,14. A phage stock prepared from such a strain can then be used to infect a recipient E. coli strain of interest, where at a low but reliable frequency the Kan cassette-containing region can be incorporated into the recipient genome by homologous recombination (Figure 2). Transductants can be selected for the growth on the Kan-containing medium. Following this, if removal of the antibiotic resistance cassette is desired, the FLP recombinase can be supplied to the transductant strain in trans. After curing the FLP-containing plasmid, which carries an ampicillin (Amp) resistance marker, Kan and Amp-sensitive clones are screened for, and the correct excision of the wild-type coding sequence and the Kan cassette are verified by colony PCR.
Here, a detailed protocol is presented, describing each of the steps in producing a knock-out E. coli strain based on the strategy outlined above. As an example, a deletion of the tamA gene is demonstrated. tamA encodes an outer membrane β-barrel protein that is a part of the Transport and Assembly Module (TAM), which is involved in the biogenesis of certain autotransporter proteins and pili15,16,17. This knock-out strain was then used to examine the effect of the tamA deletion on the biogenesis of two trimeric autotransporter adhesins (TAAs), the Yersinia adhesin YadA and the E. coli immunoglobulin (Ig)-binding TAA EibD18,19.
Protocol
1. Strains and Plasmids
- Bacterial strains
- Use the E. coli strains BW251135, JW4179 (BW25113 tamA::kan)7, BL21(DE3)20, and BL21ΔABCF21. See Table of Materials for further information.
- Bacteriophages
- Use the phage P1vir for the general transduction. Store the phage as a liquid stock with a few drops of chloroform (see step 2.2). For more information, see Table of Materials.
- Plasmids
- Use the following plasmids in this protocol: pCP2022, pIBA2-YadA23, and pEibD1024. As control plasmids, use pASK-IBA2 and pET22b (see Table of Materials).
- Growth conditions
- Propagate bacteria in a lysogeny broth (LB) medium25 with vigorous shaking (180–200 rpm) at 37 °C or 30 °C in the case of BL21ΔABCF and strains containing pCP20.
- Perform plasmid curing at 43 °C.
- For a solid medium, supplement LB with 1% agar (w/v).
- For top agar, supplement LB with 0.7% agar and 10 mM CaCl2 and autoclave the medium. Use SOC medium for the recovery after electroporation26.
- Use the following concentrations for antibiotics: 100 μg/mL for Amp and 25 μg/mL for Kan.
2. Preparing a Phage Lysate
- Infection of the donor strain
- Grow the donor strain JW4197 in 5 mL of LB medium supplemented with 10 mM CaCl2 and optionally with Kan (25 µg/mL) to an optical density at 600 nm (OD600) of ~1.0. Measure the OD600 value using a spectrophotometer.
- Make a dilution series of an existing P1 phage stock in the LB medium: recommended dilutions are between 10-3 to 10-7.
- Mix 200 μL of the bacterial suspension and 100 μL of a given phage dilution in a 15 mL centrifuge tube or equivalent. Prepare as many tubes as phage dilutions. Incubate the tubes for 20 min at 37 °C without shaking.
- Add ~3 mL of molten top agar (~50 °C) supplemented with 10 mM CaCl2 to the tubes, mix the contents thoroughly by vortexing the tubes shortly, and pour the mixtures onto prewarmed LB plates to make even layers.
- Incubate the plates overnight at 37 °C.
- Lysate preparation
- The following day choose a plate with a semi-confluent growth of phage plaques. On a semi-confluent plate, approximately half the surface area of the plate is clear (Figure 3).
- Scrape the top agar layer from such a plate using an inoculation loop or a similar tool and place the top agar in a centrifuge tube. Add 1–2 mL of LB and a drop of chloroform and vortex the tube vigorously for ~1 min. Add the chloroform in a fume hood.
- Centrifuge the tube for 15 min at 4,000 x g or faster to pellet the agar and bacterial cells.
- Move the supernatant to a fresh microcentrifuge tube, avoiding carrying over any debris from the pellet. Add 2 drops of chloroform and store the lysate at 4–10 °C. Add chloroform in a fume hood. Do not freeze the phage lysate as this will result in a significant reduction of the number of infectious particles.
- Determining the lysate titer
- Grow BW25113 in LB supplemented with 10 mM CaCl2 at 37 °C till the culture reaches an OD600 of ~1.0.
- Prepare a dilution series of the phage lysate in LB (e.g., 10-6–10-9).
NOTE: Be careful to pipet the sample from the top of the lysate to avoid transferring chloroform to the dilutions. - Mix 200 μL of the bacterial suspension and 100 μL of a given phage dilution in a 15 mL centrifuge tube or equivalent. Prepare as many tubes as phage dilutions. Incubate the tubes for 20 min at 37 °C without shaking.
- Add ~3 mL of molten top agar (~50 °C) supplemented with 10 mM CaCl2 to the tubes, mix the contents thoroughly by vortexing the tubes shortly, and pour the mixture onto prewarmed LB plates to make even layers.
- Incubate the plates overnight at 37 °C.
- The following day count the number of plaques (clear regions in the bacterial mat) for each plate and calculate the titer of the lysate using the following formula:

Example: On plates with the dilutions 10-7, 10-8, and 10-9, there are 231, 27, and 2 plaques, respectively. As 100 μL of each dilution was used for the infection, and because the titer is expressed as plaque-forming units (pfu)/mL, the plating factor is 10. These numbers are entered into the formula:
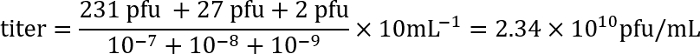
Thus, the titer is approximately 2 x 1010 infectious phage particles/mL.
3. P1 Transduction
- Preparing the recipient cells
- Grow the recipient strain BL21ΔABCF in LB supplemented to an optical density at 600 nm (OD600) of ~1.0. Use a spectrophotometer to measure the OD600 value.
- Calculate the volume of the phage lysate needed to achieve a multiplicity of infection (MOI) value of 0.5. To calculate the MOI, estimate the number of bacteria based on the OD600 of the culture. Assume that an OD600 value of 1.0 corresponds to ~109 cfu/mL. Calculate the volume required based on the known titer of the lysate.
Example (based on the titer calculated in the example after step 2.3.6):

- Add CaCl2 to the recipient strain culture to 10 mM and mix. Take 1 mL of the culture for transduction.
- Performing transduction
- Add the appropriate volume of phage lysate to 1 mL of recipient culture (including CaCl2 at 10 mM) as calculated in step 3.1.2 and mix gently.
NOTE: Be careful to pipette the sample from the top of the lysate to avoid transferring chloroform to the mix. - Statically incubate the mix for 20 min at 37 °C.
- Stop the infection by adding sodium citrate, pH 5.5, to 100 mM.
- Centrifuge the bacteria (5,000 x g for 2 min) and remove the supernatant, then resuspend them in 1 mL of fresh LB supplemented with 100 mM sodium citrate, pH 5.5.
- Wash the cells twice more as in step 3.2.4 to ensure the removal of free phages and calcium.
- Resuspend the bacteria in 1 mL of fresh LB supplemented with 100 mM sodium citrate, pH 5.5. Incubate the bacteria at 30 °C for 1 h with shaking (> 100 rpm).
- Collect the bacteria by centrifugation (5,000 x g for 2 min) and resuspend them in ~100 μL of LB with 100 mM sodium citrate, pH 5.5.
- Spread the bacteria on an LB plate supplemented with Kan at 25 µg/mL and 10 mM sodium citrate, pH 5.5, and grow the bacteria at 30 °C until colonies appear (~24 h).
- Add the appropriate volume of phage lysate to 1 mL of recipient culture (including CaCl2 at 10 mM) as calculated in step 3.1.2 and mix gently.
- Selecting the transductants
- Once colonies have grown on the selection plate, restreak them on LB + Kan for single colonies and grow at 30 °C till single colonies appear.
4. Excising the Kan cassette
- The transformation with recombination plasmid
- Make the Kan-resistant BL21ΔABCF strain electrocompetent.
- Grow the strain from a single colony in fresh LB (5 mL) supplemented with Kan (25 µg/mL) at 30 °C until the OD600 is ~0.5–0.7.
- From here on, carry out the following steps at 4 °C or on ice. Centrifuge the bacteria (5,000 x g for 10 min) and remove the supernatant.
- Wash the cell pellet with ice-cold distilled water twice by repeating the previous centrifugation step and, finally, resuspend cell pellet in 100 µL of ice-cold 10% glycerol.
- Cool down 1 mm electroporation cuvettes on ice.
- Add 1 pg of plasmid DNA (pCP20) to the cell suspension, mix the suspension gently, and transfer it to a cooled electroporation cuvette.
- Set the electroporator to 1.8 kV and electroporate the cells.
- Rescue transformed cells by adding 1 mL of SOC medium and growing them for 1 h at 30 °C.
- Plate 100 µL of the cells on LB + Amp plates and grow the cells at 30 °C overnight.
- Make the Kan-resistant BL21ΔABCF strain electrocompetent.
- Induction of the recombination
- Pick single colonies from the LB + Amp plate and inoculate fresh LB omitting all antibiotics.
- Grow the cells at the non-permissive temperature (43 °C) overnight to induce the expression of FLP recombinase.
- Selecting recombinants
- Make serial dilutions and plate 50 µL of a 105-106 dilution on non-selective plates and grow it overnight at 30 °C.
5. Verification of the Gene Deletion
- Verification of the plasmid loss and successful recombination
- Streak single colonies from the plates prepared in step 4.3.1 on LB + Kan, LB + Amp, and LB plates without antibiotics, in this order. To aid in the streaking, use a colony grid (see Supplementary File 1). Grow the plates at 30 °C until colonies appear (~24 h).
- Pick 10–20 clones that have grown on the non-selective plate but failed to grow on the selective media for further verification.
- Additional verification by colony PCR
- Perform a colony PCR with primers flanking the tamA coding sequence (see Table of Materials).
- Prepare a master PCR mix. The amount of mix needed depends on the number of colonies to be tested (see the example in Table 1). Mix the reagents on ice, add the polymerase last.
- Mix the PCR master mix thoroughly, then dispense 20 µL into tubes of a PCR strip. Pick a small amount of each colony to be screened using a sterile pipette tip and add it to a tube. Remember to include the original recipient strain for comparison. Optionally, also include the donor strain.
- Run a PCR reaction (see Table 2 for the program used).
- Prepare a 1% agarose gel.
- For a 50 mL gel, measure 0.5 g of agarose and add 50 mL of TAE buffer (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA, pH 8.0). Heat the mixture in a microwave oven until all agarose has dissolved.
- Once the agarose has dissolved, cool the solution to approximately 50 °C, and then add 5 μL of DNA staining dye. Mix the solution well and pour it into a gel tray in a casting chamber. Insert one or more rows of well combs so that there are sufficient wells for all samples, as well as molecular size markers. Allow the gel to set for 30 min.
- Once the PCR run is complete, add 4 µL of 6x DNA loading dye to each sample. Place the gel in the electrophoresis chamber and add TAE buffer till the wells are covered.
- Apply 10–15 µL from each PCR reaction to a well in the gel. Also add 5 µL of a molecular size marker. Then, run the gel for 30 min at 75 V.
- Once the run is complete, image the gel using a blue-light imager.
6. Other Techniques
- Protein expression
NOTE: The expression of test proteins (YadA and EibD) has been described in detail elsewhere23,24. This is a brief summary of the main steps.- Transform BL21ΔABCF ΔtamA with the necessary plasmids (pIBA2-YadA and pEibD10 and the corresponding control plasmids) and select for transformants on LB + Amp.
- For the protein expression, inoculate 100 mL of LB medium + Amp with 1 mL of an overnight culture of transformed bacteria and grow these at 30 °C until mid-log phase, at which time, induce the protein production with either anhydrotetracycline (at 100 ng/mL) or isopropyl thiogalactoside (at 0.5 mM).
- After 2 h of induction at 30 °C, measure the turbidity of the cultures and collect a number of cells corresponding to 50 mL at OD600 = 1.0 by centrifuging the cultures for 15 min at 4,000 x g. Wash the pellet 1x with 10 mM HEPES, pH 7.4, and then either store it at -20 °C or process it further as in step 6.2.
- Outer membrane extraction
NOTE: Outer membrane extraction is performed as described in detail by Leo et al.27. The main steps are summarized below.- Resuspend the cell pellet in 1 mL of 10 mM HEPES, pH 7.4, supplemented with 10 mM MgCl2 and MnCl2, lysozyme (0.1 mg/mL), and a pinch of DNase I.
- Lyse the cells (e.g., using a bead beater).
- Centrifuge the cells shortly (2 min at 15,600 x g) to remove cell debris and move the supernatant to a fresh tube.
- Centrifuge the cells for 30 min at 16,000 x g, after which, resuspend the pellet in 400 μL of 1% N-lauroyl sarcosine in 10 mM HEPES, pH 7.4.
- Incubate the cells with agitation for 30 min at room temperature, after which, centrifuge them for 30 min as above.
- Wash the translucent pellet 1x with 200 μL of 10 mM HEPES, pH 7.4, and then resuspend it in 30 μL of 10 mM HEPES and add 10 μL of 4x SDS-PAGE sample buffer.
- Activity assays
NOTE: Perform SDS-PAGE and activity assays for YadA and EibD as described28. The main steps are summarized below. - SDS-PAGE
- Heat the samples at 50 °C for 5 min before loading them onto a polyacrylamide gel, to avoid denaturing the proteins.
- After the separation in SDS-PAGE, transfer the proteins to a polyvinylidene difluoride (PVDF) membrane.
- After the transfer, block the membrane with 2% skimmed-milk powder in PBS.
- YadA-collagen far-western blot
- After the blocking, add bovine collagen type I diluted in blocking buffer to a concentration of 10 μg/mL and incubate the membrane for 1 h.
- Wash the membrane 2x with PBS + 0.05% Tween20 (PBS-T).
- Add the primary antibody (monoclonal anti-collagen COL-1) to the membrane, diluted 1:2,000 in blocking buffer.
- After incubating the membrane for 1 h, wash 2x as mentioned in step 6.3.2.2 and then add the secondary antibody [goat anti-mouse IgG-horseradish peroxidase (HRP) conjugate], diluted 1:10,000 in blocking buffer.
- Incubate the membrane for 1 h, then wash it 2x with PBS-T. Add an enhanced chemiluminescent substrate to the membrane according to the manufacturer’s instructions and detect the band using a CCD camera.
- EibD-IgG binding assay
- After the blocking, add a secondary antibody (goat anti-rabbit HRP), diluted 1:2,000 in blocking buffer.
- Incubate the membrane for 1 h, then wash it 2x with PBS. Perform chemiluminescent detection as mentioned in step 6.3.2.5.
Results
Generation of a tamA Knock-out of BL21ΔABCF:
The strategy outlined above has previously been used to produce a derivative strain of BL21(DE3), a standard laboratory strain used for protein production, which is optimized for outer membrane protein production and called BL21ΔABCF21. This strain lacks four genes coding for abundant outer membrane proteins and, consequently, is able to produce...
Discussion
P1 transduction is a fast, robust, and reliable method for generating gene deletions in E. coli. This is demonstrated here by transducing a tamA deletion mutant from a Keio donor strain to a BL21-derived recipient. The major stages in the transduction process are the production of the transducing lysate, the transduction itself, the excision of the Kan resistance cassette, and the verification of the knock-out by PCR. In total, the process takes approximately 1 week and requires no molecular biology met...
Disclosures
The authors have nothing to disclose.
Acknowledgements
Keio collection strains were obtained from the National BioResource Project (NIG, Japan): E. coli. We thank Dirk Linke (Department of Biosciences, University of Oslo) for his continuing support. This work was funded by the Research Council of Norway Young Researcher grant 249793 (to Jack C. Leo).
Materials
| Name | Company | Catalog Number | Comments |
| Strains | |||
| E. coli BW25113 | NIG | ME6092 | Wild-type strain of Keio collection |
| E. coli BL21(DE3) | Merck | 69450-3 | Expression strain |
| E. coli BL21DABCF | Addgene | 102270 | Derived from BL21(DE3) |
| E. coli JW4179 | NIG | JW4179-KC | tamA deletion mutant |
| P1 vir | NIG | HR16 | Generally transducing bacteriophage |
| Plasmids | |||
| pCP20 | CGSC | 14177 | conditionally replicating plasmid with FLP |
| pASK-IBA2 | IBA GmbH | 2-1301-000 | expression vector |
| pEibD10 | N/A | N/A | for production of EibD; plasmid available on request |
| pET22b+ | Merck | 69744-3 | expression vector |
| pIBA2-YadA | N/A | N/A | for production of YadA; plasmid available on request |
| Chemicals | |||
| Acetic acid | ThermoFisher | 33209 | |
| Agar | BD Bacto | 214010 | |
| Agarose | Lonza | 50004 | |
| Ampicillin | Applichem | A0839 | |
| Anhydrotetracycline | Abcam | ab145350 | |
| anti-collagen type I antibody COL-1 | Sigma | C2456 | |
| Bovine collagen type I | Sigma | C9791 | |
| Calcium chloride | Merck | 102382 | |
| Chloroform | Merck | 102445 | |
| Di-sodium hydrogen phosphate | VWR | 28029 | |
| DNA dye | Thermo | S33102 | |
| DNA molecular size marker | New England BioLabs | N3232S | |
| DNase I | Sigma | DN25 | |
| dNTP mix | New England Biolabs | N0447 | |
| ECL HRP substrate | Advansta | K-12045 | |
| EDTA | Applichem | A2937 | |
| Glycerol | VWR | 24388 | |
| goat anti-mouse IgG-HRP | Santa Cruz | sc-2005 | |
| goat anti-rabbit IgG-HRP | Agrisera | AS10668 | |
| HEPES | VWR | 30487 | |
| Isopropyl thiogalactoside | VWR | 43714 | |
| Kanamycin | Applichem | A1493 | |
| Lysozyme | Applichem | A4972 | |
| Magnesium chloride | VWR | 25108 | |
| Manganese chloride | Sigma | 221279 | |
| N-lauroyl sarcosine | Sigma | L9150 | |
| Skim milk powder | Sigma | 70166 | |
| Sodium chloride | VWR | 27808 | |
| tamA forward primer | Invitrogen | N/A | Sequence 5'-GAAAAAAGGATATTCAGGAGAAAATGTG-3' |
| tamA reverse primer | Invitrogen | N/A | Sequence 5'-TCATAATTCTGGCCCCAGACC-3' |
| Taq DNA polymerase | New England Biolabs | M0267 | |
| Tri-sodium citrate | Merck | 106448 | |
| Tryptone | VWR | 84610 | |
| Tween20 | Sigma | P1379 | |
| Yeast extract | Merck | 103753 | |
| Equipment | |||
| Agarose gel electrophoresis chamber | Hoefer | SUB13 | |
| Bead beater | Thermo | FP120A-115 | |
| CCD camera | Kodak | 4000R | |
| Electroporation cuvettes | Bio-Rad | 165-2089 | |
| Electroporation unit | Bio-Rad | 1652100 | |
| Gel imager | Nippon Genetics | GP-03LED | |
| Incubating shaker | Infors HT | Minitron | |
| Incubator | VWR | 390-0482 | |
| Microcentrifuge | Eppendorf | 5415D | |
| Microwave oven | Samsung | CM1099A | |
| PCR machine | Biometra | Tpersonal | |
| PCR strips | Axygen | PCR-0208-CP-C | |
| pH meter | Hanna Instruments | HI2211-01 | |
| PVDF membrane | ThermoFisher | 88518 | |
| SDS-PAG electrophoresis chamber | ThermoFisher | A25977 | |
| Tabletop centrifuge | Beckman Coulter | B06322 | |
| Vortex mixer | Scientific Industries | SI-0236 | |
| Water bath | GFL | D3006 | |
| Wet transfer unit | Hoefer | TE22 |
References
- Blount, Z. D. The unexhausted potential of E. coli. eLife. 4, e05826 (2015).
- Hamilton, C. M., Aldea, M., Washburn, B. K., Babitzke, P., Kushner, S. R. New method for generating deletions and gene replacements in Escherichia coli. Journal of Bacteriology. 171 (9), 4617-4622 (1989).
- Link, A. J., Phillips, D., Church, G. M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. Journal of Bacteriology. 179 (20), 6228-6237 (1997).
- Sawitzke, J. A., Thomason, L. C., Costantino, N., Bubunenko, M., Datta, S., Court, D. L. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods in Enzymology. 421, 171-199 (2007).
- Datsenko, K. A., Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 97 (12), 6640-6645 (2000).
- Muyrers, J. P., Zhang, Y., Stewart, A. F. Techniques: Recombinogenic engineering--new options for cloning and manipulating DNA. Trends in Biochemical Sciences. 26 (5), 325-331 (2001).
- Baba, T., et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology. 2, (2006).
- Yamamoto, N., et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Molecular Systems Biology. 5, 335 (2009).
- Baba, T., Huan, H. -. C., Datsenko, K., Wanner, B. L., Mori, H. The applications of systematic in-frame, single-gene knockout mutant collection of Escherichia coli K-12. Methods in Molecular Biology. 416, 183-194 (2008).
- Chattopadhyay, M. K., Tabor, C. W., Tabor, H. Polyamines are not required for aerobic growth of Escherichia coli: preparation of a strain with deletions in all of the genes for polyamine biosynthesis. Journal of Bacteriology. 191 (17), 5549-5552 (2009).
- Xie, X., Wong, W. W., Tang, Y. Improving simvastatin bioconversion in Escherichia coli by deletion of bioH. Metabolic Engineering. 9 (4), 379-386 (2007).
- Maeda, T., Sanchez-Torres, V., Wood, T. K. Escherichia coli hydrogenase 3 is a reversible enzyme possessing hydrogen uptake and synthesis activities. Applied Microbiology and Biotechnology. 76 (5), 1035-1042 (2007).
- Meehan, B. M., Landeta, C., Boyd, D., Beckwith, J. The disulfide bond formation pathway is essential for anaerobic growth of Escherichia coli. Journal of Bacteriology. 199 (16), (2017).
- Niba, E. T. E., Naka, Y., Nagase, M., Mori, H., Kitakawa, M. A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Research. 14 (6), 237-246 (2008).
- Heinz, E., et al. Conserved features in the structure, mechanism, and biogenesis of the inverse autotransporter protein family. Genome Biology and Evolution. 8 (6), 1690-1705 (2016).
- Selkrig, J., et al. Discovery of an archetypal protein transport system in bacterial outer membranes. Nature Structural & Molecular Biology. 19 (5), 506-510 (2012).
- Stubenrauch, C., et al. Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine. Nature Microbiology. 1 (7), 16064 (2016).
- Leo, J. C., Goldman, A. The immunoglobulin-binding Eib proteins from Escherichia coli are receptors for IgG Fc. Molecular Immunology. 46 (8-9), 1860-1866 (2009).
- Mühlenkamp, M., Oberhettinger, P., Leo, J. C., Linke, D., Schütz, M. S. Yersinia adhesin A (YadA) - Beauty & beast. International Journal of Medical Microbiology. 305 (2), 252-258 (2015).
- Studier, F. W., Moffatt, B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. Journal of Molecular Biology. 189 (1), 113-130 (1986).
- Meuskens, I., Michalik, M., Chauhan, N., Linke, D., Leo, J. C. A new strain collection for improved expression of outer membrane proteins. Frontiers in Cellular and Infection Microbiology. 7, 464 (2017).
- Cherepanov, P. P., Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 158 (1), 9-14 (1995).
- Grosskinsky, U., et al. A conserved glycine residue of trimeric autotransporter domains plays a key role in Yersinia adhesin A autotransport. Journal of Bacteriology. 189 (24), 9011-9019 (2007).
- Leo, J. C., et al. The structure of E. coli IgG-binding protein D suggests a general model for bending and binding in trimeric autotransporter adhesins. Structure. 19 (7), 1021-1030 (1993).
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. Journal of Bacteriology. 62 (3), 293-300 (1951).
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology. 166 (4), 557-580 (1983).
- Leo, J. C., Oberhettinger, P., Linke, D. Assessing the outer membrane insertion and folding of multimeric transmembrane β-barrel proteins. Methods in Molecular Biology. , 157-167 (2015).
- Mikula, K. M., Leo, J. C., Łyskowski, A., Kedracka-Krok, S., Pirog, A., Goldman, A. The translocation domain in trimeric autotransporter adhesins is necessary and sufficient for trimerization and autotransportation. Journal of Bacteriology. 194 (4), 827-838 (2012).
- Thomason, L. C., Costantino, N., Court, D. L., Ausubel, F. M. E. coli genome manipulation by P1 transduction. Current Protocols in Molecular Biology. , (2007).
- Franklin, N. C. Mutation in gal U gene of E. coli blocks phage P1 infection. Virology. 38 (1), 189-191 (1969).
- Jeong, H., et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). Journal of Molecular Biology. 394 (4), 644-652 (2009).
- Greene, E. C. DNA Sequence Alignment during Homologous Recombination. Journal of Biological Chemistry. 291 (22), 11572-11580 (2016).
- Watt, V. M., Ingles, C. J., Urdea, M. S., Rutter, W. J. Homology requirements for recombination in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 82 (14), 4768-4772 (1985).
- Ho, T. D., Waldor, M. K. Enterohemorrhagic Escherichia coli O157:H7 gal mutants are sensitive to bacteriophage P1 and defective in intestinal colonization. Infection and Immunity. 75 (4), 1661-1666 (2006).
- Ikeda, H., Tomizawa, J. I. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. Journal of Molecular Biology. 14 (1), 85-109 (1965).
- Murooka, Y., Harada, T. Expansion of the host range of coliphage P1 and gene transfer from enteric bacteria to other Gam-negative bacteria. Applied and Environmental Microbiology. 38 (4), 754-757 (1979).
- Goldberg, R. B., Bender, R. A., Streicher, S. L. Direct selection for P1-sensitive mutants of enteric bacteria. Journal of Bacteriology. 118 (3), 810-814 (1974).
- O'Connor, K. A., Zusman, D. R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. Journal of Bacteriology. 155 (1), 317-329 (1983).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved