A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Quantitative Fluorescence In Situ Hybridization (FISH) and Immunofluorescence (IF) of Specific Gene Products in KSHV-Infected Cells
In This Article
Summary
We describe a protocol utilizing fluorescence in situ hybridization (FISH) to visualize multiple herpesviral RNAs within lytically infected human cells, either in suspension or adherent. This protocol includes quantification of fluorescence producing a nucleocytoplasmic ratio and can be extended for simultaneous visualization of host and viral proteins with immunofluorescence (IF).
Abstract
Mechanistic insight arrives from careful study and quantification of specific RNAs and proteins. The relative locations of these biomolecules throughout the cell at specific times can be captured with fluorescence in situ hybridization (FISH) and immunofluorescence (IF). During lytic herpesvirus infection, the virus hijacks the host cell to preferentially express viral genes, causing changes in cell morphology and behavior of biomolecules. Lytic activities are centered in nuclear factories, termed viral replication compartments, which are discernable only with FISH and IF. Here we describe an adaptable protocol of RNA FISH and IF techniques for Kaposi's sarcoma-associated herpesvirus (KSHV)-infected cells, both adherent and in suspension. The method includes steps for the development of specific anti-sense oligonucleotides, double RNA FISH, RNA FISH with IF, and quantitative calculations of fluorescence intensities. This protocol has been successfully applied to multiple cell types, uninfected cells, latent cells, lytic cells, time-courses, and cells treated with inhibitors to analyze the spatiotemporal activities of specific RNAs and proteins from both the human host and KSHV.
Introduction
In their lytic (active) phase, herpesviruses hijack the host cell, causing changes in cell morphology and localization of biological molecules, to produce virions. The base of operations is the nucleus, where the double-stranded DNA viral genome is replicated and packaged into a protein shell, called a capsid1. To begin, the virus expresses its own proteins, hijacking host machinery and preventing expression of non-essential host genes, a process termed the host shutoff effect. The majority of this activity is localized to specific 4′,6-diamidino-2-phenylindole (DAPI)-free nuclear regions called viral replication compartments, comprised of both host and viral proteins, RNAs, and viral DNA2. The cell is overhauled to provide space and resources for the replication compartments and thus assembly of viral capsids. Once the capsid exits the nucleus, how the capsid is enveloped in the cytoplasm to produce a membrane-bound viral particle, also known as a virion, is unclear. Understanding of the localization and spatial shifts of both host and viral biomolecules during the lytic phase provides deeper mechanistic insight into the arrangement of the replication compartment, host shutoff effect, the virion-egress pathway, and other processes related to herpesviral infection and replication.
Currently the best method to detect and study these changes is the visualization of proteins and RNAs in infected cells with immunofluorescence (IF) and fluorescent in situ hybridization (FISH), respectively. Use of a time-course with these techniques reveals the localization of biomolecules at key points of the lytic phase or simply, spatiotemporal data. FISH and IF complement other biochemical techniques, such as inhibition of a cellular process (e.g., inhibition of viral DNA replication), RT-qPCR (real-time polymerase chain reaction), RNA sequencing, Northern blots, mass spectrometry, Western blotting, and analysis of viral DNA production, that may provide a more global picture of cellular activities.
We developed RNA FISH strategies to examine the RNA products from specific genes and a computational analysis that quantitatively calculates the nucleocytoplasmic ratio of a specific gene product. The sample preparation, modified from earlier publications by Steitz and colleagues3,4, is relatively easy and can be used for both adherent and suspended cells. The protocol is also adaptable for simultaneous use of multiple RNA FISH strategies (double RNA FISH) or RNA FISH with IF strategies. Development of a specific FISH strategy is challenging, but suggestions to improve success are outlined. The data analysis described here is quantitative if fluorescent beads and strong markers of compartment boundaries are used and offers additional insight into the micrographs, insight that removes observation bias. The detailed protocol is designed for both latent and lytic cells infected by Kaposi's sarcoma-associated herpesvirus (KSHV) and can be used with uninfected cells or cells infected by other herpesviruses5. The methods of quantitation are applicable to studies on nucleocytoplasmic shifts or relocalization between subcellular compartments in most cells.
Protocol
1. Design of fluorescence in situ (FISH) anti-sense oligonucleotides to detect a specific herpesviral transcript
- Select 25 to 40 nt segments from the sequence of RNA of interest and convert to be anti-sense. A successful FISH strategy may contain from one up to ten or more different anti-sense oligonucleotides. When selecting sequences, consider the following:
- If the RNA of interest contains a unique repeat region, then capitalize on this feature and design an anti-sense oligonucleotide to target the repeat sequence.
NOTE: Tycowski and colleagues5 provide an example of this strategy with rhesus rhadinovirus (RRV) polyadenylated nuclear (PAN) RNA. - If the RNA of interest contains a known protein-binding site or stem-loop structure, design oligonucleotides that avoid these regions.
- Depending on the goals of the experiments, consider the intronic sequence and whether or not to design an anti-sense oligonucleotide for it.
- If the RNA of interest contains a unique repeat region, then capitalize on this feature and design an anti-sense oligonucleotide to target the repeat sequence.
- Perform simple computational analyses on the selected anti-sense sequences to ensure binding specificity and reduce aggregation of the anti-sense oligonucleotide.
- The sequences must be approximately 50% GC-rich (high guanine and cytosine content) and have a melting temperature in the range of 60 to 70 °C.
- Use a sequence analyzer tool to select sequences that do not self-dimerize or form hairpins with melting temperatures above 37 °C, the hybridization temperature.
- Perform a NCBI BLASTn (National Center for Biotechnology Information Basic Local Alignment Search Tool for nucleotide alignments) search of the selected sequences against both the host and viral transcriptomes using the 'somewhat similar' setting. This search will identify unique anti-sense oligonucleotides that will not likely bind to other host or viral transcripts.
NOTE: If transcriptomes are not available, perform the BLAST search with the genomic sequences. It is ideal if the searches are performed on the sequences from the virus isolated from the infected cells used in the experiment because wild strains tend to diversify and contain a combination of sequences from different lab strains.
- Order purified DNA oligonucleotides corresponding to the anti-sense sequence that has been verified computationally to be unique and likely to bind to the target RNA. No special modifications need to be introduced into the oligonucleotides.
- Test the designed anti-sense oligonucleotides for binding specificity by FISH and Northern blot.
- Using an uninfected cell-line from the same host species (e.g., 293T) and ideally the same cell type, conduct a transfection with a plasmid expressing the RNA of interest from a robust promoter (CMV, cytomegalovirus) and one with the empty vector (e.g., pcDNA3). Use a positive control for transfection such as co-transfection with a GFP (green fluorescent protein) plasmid (e.g., pmaxGFP) or the vector containing GFP.
NOTE: It is important to avoid cell-lines that have been immortalized using the herpesvirus Epstein-Barr virus (EBV) since there are sequence similarities between herpesviruses. - Perform FISH as described in section 3 on both sets of cells with the anti-sense oligonucleotides as described in this protocol. Deduce the successful candidates by comparing FISH experiments with individual, pairs, or sets of the anti-sense oligonucleotides. Use a positive control for the FISH protocol such U2 snRNA (small nuclear RNA) FISH, present at 500,000 copies per human cell nucleus6 (Table 1).
- The fluorescent signal should be specific and strong in the cell containing the RNA of interest. Design additional anti-sense oligonucleotides to strengthen the signal and remove anti-sense oligonucleotides that bind nonspecifically from consideration. Signal strength must be above background and autofluorescence.
- Test binding specificity by Northern blot.
- Using an uninfected cell-line from the same host species (e.g., 293T) and ideally the same cell type, conduct a transfection with a plasmid expressing the RNA of interest from a robust promoter (CMV, cytomegalovirus) and one with the empty vector (e.g., pcDNA3). Use a positive control for transfection such as co-transfection with a GFP (green fluorescent protein) plasmid (e.g., pmaxGFP) or the vector containing GFP.
2. Oligonucleotide and cell preparation
- Following the manufacturer's instructions, use terminal transferase to label the anti-sense oligonucleotides with dioxigenin(DIG)-dUTP or, if strongly binding, directly with a fluorescent nucleotide like Alexa Fluor 594-5-dUTP. After labeling, additional purification is not necessary. Store labeled oligonucleotides at -20 °C up to several years and in tin foil if directly labeled to prevent photobleaching.
CAUTION: The labeling solution contains a toxic material, potassium cacodylate. Handle labeling reactions with gloves.
NOTE: To conserve resources, several different anti-sense oligonucleotides can be labeled in one reaction. This protocol utilizes 3'-end labeling. Internal labeling is challenging because the chemical group (e.g., DIG or the fluorophore) gets caught or is unable to enter the active site of a DNA polymerase. An experiment with two different RNAs can be performed using directly-labeled anti-sense oligonucleotides and anti-DIG immunofluorescence with a different fluorophore (e.g., FITC (fluorescein) or Alexa Fluor 488 with Alexa Fluor 594). - Adhere the cells to the eight-chamber slides.
NOTE: Eight-chamber slides allow several simultaneous experiments while minimizing precious resources like antibodies. An alternative to eight-chamber slides is a six-well tissue culture plate with standard coverslips (22 mm x 22 mm) that are both sterile. A similar arrangement is possible with circular coverslips and a 24-well tissue culture plate. For both, increase the volumes mentioned in this protocol by 10-15x (e.g., 1.75 mL hybridization solution), and 4x (e.g., 600 µL hybridization solution) respectively.- For adherent lytic cells, use 1x trypsin/PBS at 37 °C and 5% CO2 for 10 min to suspend cells and dilute to 60% confluency.
NOTE: Adherent cell lines used in FISH experiments included 293T, iSLK.2197, and iSLK-BAC36 cells8. - Apply 200 µL of cell suspension to each chamber of the sterile eight-chambered slides and allow seed growth for 12-24 h at 37 °C and 5% CO2. Adjust as necessary for slow- or fast-growing cells and for cells that are easily damaged by trypsin.
NOTE: The objective is to have evenly spaced cells firmly attached to the slide. Consider inducing lytic phase after adhesion if the lytic cells are fragile. Conclusions drawn from experiments with iSLK cells are limited9. - For lytic suspension-cells, pre-treat eight-chamber slides with 1:10 poly L-lysine for 5 min under the tissue culture hood. Then leave the slides to dry overnight at room temperature or 1 h at 65 °C. Incubate 800 µL of lytic cells at a concentration of 1 x 106 cells/mL with the chambered slides for 30 min to 1 h at 37 °C and 5% CO2.
NOTE: Suspension-cells will settle in a monolayer, sticking to the poly L-lysine and thus excess cells are not a concern in comparison to adherent cells. Lytic cells that form grape clusters, if possible, should be separated by gentle vortexing or chemical means. Admittedly, the authors have not had much success with such recommendations in the case of lytic BJAB-RRV-GFP cells. If suspension-cells do not adhere well, consider increasing either the time or concentration of the poly L-lysine incubation.
- For adherent lytic cells, use 1x trypsin/PBS at 37 °C and 5% CO2 for 10 min to suspend cells and dilute to 60% confluency.
3. Fixation, Immunofluorescence (Optional), Hybridization, and Visualization of Viral RNAs
- Remove media and excess cells. Throughout this protocol, use vacuum suction to remove solutions and gentle micropipetting to add solutions.
NOTE: The strength of a vacuum can be reduced by placing a 200 µL micropipette tip over the glass Pasteur pipette. Replace the micropipette tip between wash steps to prevent contamination. Each wash step must be performed quickly because it is imperative that the cells never dry out. - Immediately, fix the cells with pre-chilled 4% formaldehyde/PBS (phosphate-buffered saline) on ice for 30 min. Wash the cells three times with 200 µL 1x PBS cooled to 4 °C and incubate for 5 min at room temperature or on ice.
- Permeabilize the fixed cells with 200 µL of pre-chilled 0.5% Triton-X/PBS (phosphate buffered saline) for 10 min on ice or 750 µL of pre-chilled 70% ethanol at 4 °C for 1 h (min) to 7 d (max).
NOTE: Collect protein, total RNA, and genomic DNA samples at the point of fixation to ensure consistency between images and biochemical assays. All washes throughout this protocol are performed in the same manner unless otherwise specified. 70% ethanol loosens the glue between the chambers and the slide, which eases later separation, and also provides a significant pause in the protocol. Nonetheless, use paraffin film around the chamber slide to reduce evaporation and check the level of the ethanol in each chamber about every 8 h. 70% ethanol also flattens the cells, making a crisper image, while Triton-X does not dehydrate the cells and change the dimensions of the cell.
- Permeabilize the fixed cells with 200 µL of pre-chilled 0.5% Triton-X/PBS (phosphate buffered saline) for 10 min on ice or 750 µL of pre-chilled 70% ethanol at 4 °C for 1 h (min) to 7 d (max).
- Remove chambers carefully to prevent cracking the slide. If the experiment includes immunofluorescence (IF) of a viral or host protein with a polyclonal primary antibody, perform the IF as described below before proceeding to RNA FISH. If the immunofluorescence uses a monoclonal primary antibody, then perform immunofluorescence as described in step 3.3.1 after step 3.11.
NOTE: Use a fresh removal device or one with very little leftover adhesive provided by the manufacturer and gently ease the chambers off to prevent the slide from cracking. Using 70% ethanol as the permeabilizing reagent for 4 h greatly reduces the likelihood of cracking. In the case of a crack, continue the protocol on chambers not affected by the crack and be mindful of the higher oxidation rate of imperfectly sealed slides (i.e. decreased storage life).- Rinse cells with pre-chilled 1x PBS and block with pre-chilled 4% BSA (bovine serum albumin)/1x PBS for 30 min at 4 °C.
NOTE: The use of BSA throughout this protocol limits nonspecific labeling. - Remove blocking solution and incubate the cells with 1:200 or another polyclonal primary antibody in 0.1% BSA/1x PBS for 1 h at 4 °C. Then wash three times with 1x PBS.
NOTE: An antibody10 for detection of SSB/ORF6 (viral single-stranded DNA binding protein) was used at 1:200 dilution. - Incubate the cells with a secondary antibody with fluorophore compatible with the FISH-detecting antibody for 1 h at 4 °C. Wash three times with 1x PBS. Then fix with 4% formaldehyde/1x PBS for 10-15 min and permeabilize with either Triton-X or 70% ethanol as previously described before proceeding to FISH. Cover slide with tin foil to preserve fluorescent signal and prevent photobleaching.
- Rinse cells with pre-chilled 1x PBS and block with pre-chilled 4% BSA (bovine serum albumin)/1x PBS for 30 min at 4 °C.
- Wash the cells with 2x SSC (saline sodium citrate) once and then apply 45 µL of hybridization solution consisting of 50% formamide, 10% dextran sulfate, 2x SSC, 0.1% BSA, 500 µg/mL salmon sperm DNA, 125 µg/mL E. coli tRNA, and 1 mM vanadyl ribonucleoside complexes. Incubate for 1 h at 37 °C in a humidity chamber that can be a 150 mm Petri dish with moistened sterile wipes.
NOTE: Prepare fresh hybridization solution at least an hour before use. Dissolve the dextran sulfate in water first, vortexing frequently and incubating in a 37 °C water bath. - Calculate to have a suggested concentration of 25 µM oligonucleotides in 35-uL hybridization solution per chamber. Adjust concentration of anti-sense oligonucleotide as needed. Add distilled water to the oligonucleotides to bring the denaturation volume to 10 µL.
NOTE: Following the labeling reaction, the oligonucleotides are stored in the quenched solution containing 0.18 M potassium cacodylate, 23 mM Tris-HCl, 0.23 mg/mL BSA, 4.5 mM CoCl2, 18 mM EDTA, 2.7 mM K-phosphate, and 6.8 mM KCl, 45 µM 2-Mercaptoethanol, 0.02% Triton X-100, and 2% glycerol. The concentrations are high enough that dilution with water will bring the denaturation solution to concentrations near to 1x TE (10 mM Tris-HCl and 1 mM EDTA), a standard oligonucleotide denaturation buffer. - Denature the DIG- and/or Alexa Fluor 594-labeled oligonucleotides at 95 °C for 5 min. Then add 35 µL fresh hybridization solution per intended chamber to the denatured oligonucleotides. If performing double FISH, both sets of anti-sense oligonucleotides may be denatured and hybridized together.
- Remove the pre-hybridization solution and then add hybridization solution containing the labeled oligonucleotides to the cells. Incubate overnight in the humidity chamber at 37 °C with tin foil to protect the fluorophore-labeled oligonucleotides.
NOTE: Incubation should be at least 10 h and not more than 24 h. - The next day, wash the cells twice with 2x SSC for 10 min at 37 °C and then twice with 1x SSC for 10 min at 25 °C.
- Fix the cells with pre-chilled 4% formaldehyde/1x PBS for 10-15 min on ice. Then wash the cells with PBS three times and permeabilize for 1 h with pre-chilled 70% ethanol or for 10 min with pre-chilled 0.5% Triton-X/1x PBS at 4 °C.
- Incubate the cells with 1:200 anti-DIG FITC in pre-chilled 0.1% BSA/1x PBS for 1 h at 4 °C. Remove the antibody solution and wash three times with 1x PBS.
- Fix with pre-chilled 4% formaldehyde/1x PBS for 10-15 min at 4 °C and then wash three times with 1x PBS. If performing immunofluorescence for a host or viral protein with a monoclonal primary antibody, permeabilize the cells and then perform the IF protocol outlined in step 3.3.1. Otherwise proceed to the DAPI staining.
- Incubate the cells with 0.4 µg/mL DAPI in pre-chilled 0.5% Triton-X/1x PBS for 15 min on ice and then wash three times with 1x PBS.
- Mount slides with fluorescent beads (optional) and a mounting medium. Then seal the coverslip to the slide with clear nail polish.
- Using a confocal microscope, collect images of the samples within an hour to a week of performing the protocol at 630x magnification. Apply multiple coats of nail polish to seal the cover slip and to prolong fluorophore life by reducing the rate of oxidation.
NOTE: Do not use a DAPI-containing mounting medium. When collecting the images, include the scale bar on each image for later quantification. Fluorescent beads serve as controls of fluorescence intensity between slides and sample preparations11. Acquire images at the midsection of the cell for two-dimensional (2D) quantification in step 4.
4. Quantification of FISH and IF images to highlight subcellular localization and to determine nucleocytoplasmic ratio of fluorescence
- Perform image analysis on an assembled stack of the various fluorescent-stained and merged images to ensure consistency. Set the scale of the image analysis software using the scale bar included when the images were collected.
- To quantify fluorescence intensity across several channels and in reference to the nuclear DAPI stain, use a line tool and a plot-profile function. Then indicate the line permanently on a copy of the image using markers that do not obstruct or influence the viewer's judgment.
- Establish criteria to guide where the line is drawn such as a trace that captures a diversity of topographical features, peaks and valleys, along a central axis or a line that does not traverse supersaturated areas.
NOTE: These line traces depict raw fluorescence in a cell and thus are limited to comparisons of the locations of a stain, not intensity. To compare intensities of the same stain between slides, treatments, or preparations, add a fluorescent bead to the slide as an internal control during step 3.13. The fluorescent bead must be added during the mounting process and detected with the same settings on the excitation laser and photomultiplier tube (confocal).
- Establish criteria to guide where the line is drawn such as a trace that captures a diversity of topographical features, peaks and valleys, along a central axis or a line that does not traverse supersaturated areas.
- To quantify a shift in subcellular localization, calculate nucleocytoplasmic ratios of cells undergoing different treatments.
- Measure the area and raw fluorescence intensity of both the nucleus and cytoplasm using the nuclear DAPI stain to set the inner boundary. Include nuclear and cytoplasmic controls such as a nuclear RNA (e.g., KSHV PAN RNA) and cytoplasmic RNA (e.g., host GAPDH mRNA). Moreover, calculate background intensity for three cell-like areas and average the values per pixel or µm2.
NOTE: Intensity values tend to lack units and so the term 'units' is used. - Normalize both nuclear and cellular raw intensity values by first determining the average background for the same area and then subtracting that individualized value from the raw intensity of the area.
- For example, a nucleus of a lytic B-cell has an area of 133.4 µm2 and a raw intensity of 75976 units while the background intensity for the same fluorescent signal was determined to be 0.67 units per µm2. The normalized nuclear intensity would be
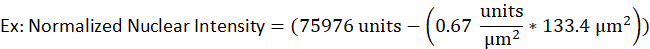
- For example, a nucleus of a lytic B-cell has an area of 133.4 µm2 and a raw intensity of 75976 units while the background intensity for the same fluorescent signal was determined to be 0.67 units per µm2. The normalized nuclear intensity would be
- Enter the values in the following equation.

NOTE: This calculation controls for changes in subcellular area. Lytic induction and drug treatments can enlarge the nucleus or change the size of the cell, respectively. - To interpret the results, create a box whisker plot. An equal distribution of the fluorescent signal would be close to zero, whereas a nuclear distribution would favor a positive ratio value and a cytoplasmic distribution would trend toward a negative ratio value.
- Measure the area and raw fluorescence intensity of both the nucleus and cytoplasm using the nuclear DAPI stain to set the inner boundary. Include nuclear and cytoplasmic controls such as a nuclear RNA (e.g., KSHV PAN RNA) and cytoplasmic RNA (e.g., host GAPDH mRNA). Moreover, calculate background intensity for three cell-like areas and average the values per pixel or µm2.
Results
The FISH and IF methods detailed in this manuscript are shown in Figure 1 along with the quantification of results by line traces of fluorescent intensity. The results presented here are semi-quantitative and offer insight into localization, rather than into comparisons between intensities of different fluorescent stains because experiments did not include a fluorescent bead in the slide preparation. Figure 1 also reveals that th...
Discussion
The protocol described in this report can be adapted to different cell types and includes steps for double RNA FISH and RNA FISH with IF using both monoclonal and polyclonal primary antibodies. Although prepared slides are typically imaged with a confocal microscope, imaging can be performed with a STED (stimulated emission depletion) microscope after modifications of increased antibody concentration and a different mounting medium. For enhanced analysis of individual cells, samples prepared with this protocol may also b...
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
We thank Jonathan Rodenfels, Kazimierz Tycowski, and Johanna B. Withers for advice on data analysis. We also thank G. Hayward for the anti-SSB antibody. This work was supported by grants T32GM007223 and T32AI055403 from the National Institutes of Health (to TKV) and NIH grant (CA16038) (to JAS). JAS is an investigator of the Howard Hughes Medical Institute. Figures 1-3 and Table 1 were reproduced with permission from the American Society for Microbiology under a Creative Commons Attribution license from the following publication: Vallery, T. K., Withers, J. B., Andoh, J. A., Steitz, J. A. Kaposi's Sarcoma-Associated Herpesvirus mRNA Accumulation in Nuclear Foci Is Influenced by Viral DNA Replication and Viral Noncoding Polyadenylated Nuclear RNA. Journal of Virology. 92 (13), doi:10.1128/JVI.00220-18, (2018).
Materials
| Name | Company | Catalog Number | Comments |
| AlexaFluor594-5-dUTP | Life Technologies | C1100 | |
| anti-DIG FITC | Jackson Lab Immunologicals | 200-092-156 | |
| Anti-Rabbit Secondary AlexaFluor594 Monoclonal Antibody | Invitrogen | A-11037 | Goat |
| Anti-SSB Antibody | N/A | N/A | Ref. Chiou et al. 2002 |
| BLASTn | NIH NCBI | N/A | Free Sequence Alignment Software |
| Dextran Sulfate | Sigma Aldrich | D8906 | Molecular Biology Grade |
| DIG-Oligonucleotide Tailing Kit | Sigma Roche | #03353583910 | 2nd Gen |
| Eight-Chamber Slides | Nunc Lab Tek II | #154453 | Blue seal promotes surface tension but separation by clear gel is also available. |
| Formamide | Sigma Aldrich | F9037 | Molecular Biology Grade |
| GAPDH Probes | Stellaris | SMF-2019-1 | Compatible with protocol, Quasar 670 |
| ImageJ | NIH, Bethesda, MD | N/A | Free Image Analysis Software, [http:rsb.info.nih.gov/ij/] |
| OligoAnalyzer | IDT | N/A | Free Oligonucleotide Analyzer |
| pcDNA3 | Invitrogen | A-150228 | |
| pmaxGFP | Amaxa | VDF-1012 | |
| Poly L-Lysine | Sigma Aldrich | P8920 | |
| Terminal Transferase | Sigma Roche | #003333574001 | |
| Vanadyl Ribonucleoside Complexes | NEB | S1402S | |
| Vectashield | Vector Laboratories, Inc. | H-1000 | DAPI within the mounting media scatters the light and reduces contrast. |
References
- Amen, M. A., Griffiths, A. Packaging of Non-Coding RNAs into Herpesvirus Virions: Comparisons to Coding RNAs. Frontiers in Genetics. 2, 81 (2011).
- Schmid, M., Speiseder, T., Dobner, T., Gonzalez, R. A. DNA virus replication compartments. Journal of Virology. 88 (3), 1404-1420 (2014).
- Pawlicki, J. M., Steitz, J. A. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. Journal of Cell Biology. 182 (1), 61-76 (2008).
- Borah, S., Darricarrere, N., Darnell, A., Myoung, J., Steitz, J. A. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. Public Library of Science Pathogens. 7 (10), e1002300 (2011).
- Tycowski, K. T., Shu, M. D., Borah, S., Shi, M., Steitz, J. A. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Reports. 2 (1), 26-32 (2012).
- Weinberg, R. A., Penman, S. Small molecular weight monodisperse nuclear RNA. Journal of Molecular Biology. 38 (3), 289-304 (1968).
- Myoung, J., Ganem, D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. Journal of Virology Methods. 174 (1-2), 12-21 (2011).
- Brulois, K. F., et al. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. Journal of Virology. 86 (18), 9708-9720 (2012).
- Sturzl, M., Gaus, D., Dirks, W. G., Ganem, D., Jochmann, R. Kaposi's sarcoma-derived cell line SLK is not of endothelial origin, but is a contaminant from a known renal carcinoma cell line. International Journal of Cancer. 132 (8), 1954-1958 (2013).
- Chiou, C. J., et al. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi's sarcoma-associated herpesvirus. Journal of Virology. 76 (7), 3421-3439 (2002).
- Cole, R. W., Jinadasa, T., Brown, C. M. Measuring and interpreting point spread functions to determine confocal microscope resolution and ensure quality control. Nature Protocols. 6 (12), 1929-1941 (2011).
- Nakamura, H., et al. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. Journal of Virology. 77 (7), 4205-4220 (2003).
- Majerciak, V., Yamanegi, K., Zheng, Z. M. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. Journal of Virology. 80 (24), 11968-11981 (2006).
- Sun, R., Lin, S. F., Gradoville, L., Miller, G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proceedings of the National Academy Sciences U S A. 93 (21), 11883-11888 (1996).
- Vallery, T. K., Withers, J. B., Andoh, J. A., Steitz, J. A. Kaposi's Sarcoma-Associated Herpesvirus mRNA Accumulation in Nuclear Foci Is Influenced by Viral DNA Replication and Viral Noncoding Polyadenylated Nuclear RNA. Journal of Virology. 92 (13), (2018).
- Borah, S., Nichols, L. A., Hassman, L. M., Kedes, D. H., Steitz, J. A. Tracking expression and subcellular localization of RNA and protein species using high-throughput single cell imaging flow cytometry. RNA. 18 (8), 1573-1579 (2012).
- Bruce, A. G., et al. Quantitative Analysis of the KSHV Transcriptome Following Primary Infection of Blood and Lymphatic Endothelial Cells. Pathogens. 6 (1), (2017).
- Chen, C. P., et al. Kaposi's Sarcoma-Associated Herpesvirus Hijacks RNA Polymerase II To Create a Viral Transcriptional Factory. Journal of Virology. 91 (11), (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved