A subscription to JoVE is required to view this content. Sign in or start your free trial.
Methods Article
Covalent Labeling with Diethylpyrocarbonate for Studying Protein Higher-Order Structure by Mass Spectrometry
In This Article
Summary
The experimental procedures for performing diethylpyrocarbonate-based covalent labeling with mass spectrometric detection are described. Diethylpyrocarbonate is simply mixed with the protein or protein complex of interest, leading to the modification of solvent accessible amino acid residues. The modified residues can be identified after proteolytic digestion and liquid chromatography/mass spectrometry analysis.
Abstract
Characterizing a protein's higher-order structure is essential for understanding its function. Mass spectrometry (MS) has emerged as a powerful tool for this purpose, especially for protein systems that are difficult to study by traditional methods. To study a protein's structure by MS, specific chemical reactions are performed in solution that encode a protein's structural information into its mass. One particularly effective approach is to use reagents that covalently modify solvent accessible amino acid side chains. These reactions lead to mass increases that can be localized with residue-level resolution when combined with proteolytic digestion and tandem mass spectrometry. Here, we describe the protocols associated with use of diethylpyrocarbonate (DEPC) as a covalent labeling reagent together with MS detection. DEPC is a highly electrophilic molecule capable of labeling up to 30% of the residues in the average protein, thereby providing excellent structural resolution. DEPC has been successfully used together with MS to obtain structural information for small single-domain proteins, such as β2-microglobulin, to large multi-domain proteins, such as monoclonal antibodies.
Introduction
Proteins are essential biomolecules in virtually every physiological process. The variety of functions that proteins perform are possible because of the structures they adopt and the interactions that they have with other biomolecules. To understand protein function at a deeper level, biochemical and biophysical tools are needed to elucidate these important structural features and interactions. Traditionally, X-ray crystallography, cryogenic electron microscopy, and nuclear magnetic resonance (NMR) spectroscopy have provided the desired atomic-level detail to reveal protein structure. However, numerous protein systems cannot be interrogated by these techniques because of poor crystallization behavior, limited protein availability, excessive sample heterogeneity, or molecular weight limitations. Consequently, newer analysis methods have emerged that overcome these limitations. Among the emerging techniques that can provide protein structural information is mass spectrometry.
Mass spectrometry (MS) measures a molecule's mass-to-charge (m/z) ratio, so protein higher-order structural information must be obtained by encoding the desired structural information into the mass of the protein. Several approaches to encode this information have been developed, including hydrogen-deuterium exchange (HDX)1,2,3,4, chemical crosslinking (XL)5,6, and covalent labeling (CL)7,8,9,10. In HDX, backbone amide hydrogens are exchanged by slightly more massive deuteriums at rates that depend on solvent accessibility and H-bonding extent. The extent of HDX can be localized by rapidly digesting the protein into peptide fragments before separating and measuring these fragments by the mass spectrometer or by dissociating the protein in a top-down experiment. Determining the rate of exchange provides further insight into protein dynamics. HDX has proven to be a valuable tool for characterizing protein structure despite challenges associated with back exchange and the need for specialized equipment to maximize reproducibility. In XL-MS, proteins are reacted with bi-functional reagents that covalently link adjacent residue side chains within a given protein or between two proteins. In doing so, XL-MS can provide distance constraints that can be used to characterize protein structure. The regions of the protein that are cross-linked can be identified by proteolytic digestion followed by liquid chromatography (LC)-MS analysis. While XL-MS is a versatile tool that has been used to study a variety of protein complexes, including inside cells, identification of the XL products is challenging and requires specialized software.
CL-MS has emerged recently as a complementary and sometimes alternative MS-based tool to study protein structure and interactions. In CL-MS, a protein or protein complex is covalently modified with a mono-functional reagent that can react with solvent-exposed side chains (Figure 1). By comparing the modification extents of a protein or protein complex under different conditions, conformation changes, binding sites, and protein-protein interfaces can be revealed. After the CL reaction, site-specific information, often at the single amino-acid level, can be obtained using typical bottom-up proteomics workflows in which proteins are proteolytically digested, peptide fragments are separated by LC, and modified sites are identified using tandem MS (MS/MS). The rich history of bioconjugate chemistry has made numerous reagents available for CL-MS experiments. CL reagents fall into two general categories: (i) specific and (ii) non-specific. Specific reagents react with a single functional group (e.g., free amines)8,10 and are easy to implement, but they tend to provide limited structural information. Non-specific reagents react with a wide range of side chains, but often require specialized equipment such as lasers or synchrotron sources to produce these highly reactive species. Hydroxyl radicals are the most commonly used non-specific reagent, having been applied in hydroxyl radical footprinting (HRF)7,11,12,13 experiments to study a wide range of proteins and protein complexes under a variety of conditions.
Our research group has successfully used another relatively non-specific reagent called diethylpyrocarbonate (DEPC) to study protein structure and interactions in the context of CL-MS experiments14,15,16,17,18,19,20,21,22,23,24,25. DEPC offers the simplicity of specific labeling reagents (i.e., no specialized equipment is necessary to perform the reactions), while reacting with up to 30% of amino acids in the average protein. As a highly electrophilic reagent, DEPC is capable of reacting with the N-terminus and the nucleophilic side chains of cysteine, histidine, lysine, tyrosine, serine, and threonine residues. Typically, a single product of these reactions is generated, resulting in a mass increase of 72.02 Da. This single type of product contrasts with the up to 55 different products that can be produced when proteins react with hydroxyl radicals7. Such simple chemistry facilitates identification of labeled sites.
Here, we provide protocols for using DEPC-based CL-MS to study protein structure and interactions. Details associated with reagent preparation, DEPC-protein reactions, protein digestion conditions, LC-MS and MS/MS parameters, and data analysis are described. We also demonstrate the utility of DEPC labeling by providing example results from protein-metal, protein-ligand, and protein-protein interactions as well as proteins undergoing structural changes upon heating.
Protocol
1. Protein and reagent preparation
NOTE: This protocol includes an example workflow for labeling a protein with DEPC. Some conditions and reagent concentrations listed may vary based on the protein of choice.
- Prepare all reagent solutions in 1.5 mL microcentrifuge tubes.
- Prepare a protein solution of desired concentration, usually in the range of tens of µM, in a 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer at pH 7.4. Alternatively, prepare a buffer-exchange existing protein solution in 10 mM pH 7.4 MOPS if the sample contains a nucleophilic buffer that would be reactive with DEPC. Other buffers (e.g., phosphate buffered saline) can also be used, as long as they do not have nucleophilic functional groups.
- Prepare a 100 mM DEPC solution in dry acetonitrile (ACN) by pipetting 1.45 µL of the stock 6.9 M DEPC solution into 98.55 µL of the ACN.
NOTE: There is no one set of concentrations that will work with every protein, though the optimal concentrations can be estimated based on the number of His and Lys residues23. For example, with 50 µL of a 50 µM β2-microglobulin solution, react the protein with 0.2 µL of the 100 mM DEPC for a final DEPC concentration of 200 µM (equal to 4x the protein concentration) to ensure the desired molar ratio using this general example (Table 1). DEPC labeling is a 2nd order reaction, so changing either the concentration of protein or DEPC in the reaction mixture will change the labeling rate. - Prepare 1 M imidazole solution by weighing out 10 mg of imidazole and dissolving in 146.9 µL of HPLC-grade water.
2. Covalent labeling of intact protein
- Set water bath temperature to 37 °C and wait for the bath to reach a stable temperature.
NOTE: Reagent concentrations and volumes for an example labeling protocol can be found in Table 1. - In a new microcentrifuge tube, mix MOPS buffer and protein solution in volumes listed in Table 1.
- To the protein and buffer add 0.2 µL of the DEPC solution, making sure to properly mix the resulting solution, and then place the tube containing the reaction mixture into the 37 °C water bath for 1 minute.
NOTE: The volume of ACN added should not exceed 1% of the total reaction volume to avoid perturbation of the protein's structure during the labeling reaction. Reaction time is up to the user, although a 1 minute reaction under the example conditions minimizes overlabeling and the potential hydrolysis of DEPC14. - After 1 minute, remove the tube containing the reaction mixture from the water bath and quench the reaction with 1 µL of the 1 M imidazole solution to scavenge the remaining unreacted DEPC.
NOTE: The final concentration of imidazole in the reaction mixture should equal 50x the concentration of DEPC in the reaction mixture. This will ensure that remaining unreacted DEPC is scavenged.
3. Preparation of protein digest for bottom-up LC-MS
NOTE: Choose digestion conditions that are amenable to the protein of interest. Common steps involve unfolding the protein and reducing and alkylating any disulfide bonds.
- Unfold the protein by adding an appropriate unfolding reagent to the reaction mixture.
NOTE: Common unfolding agents include ACN, urea, and guanidine hydrochloride (GuHCl). - Prepare solutions of Tris(2-carboxyethyl)phosphine (TCEP) and iodoacetamide (IAM) by weighing out 5 mg of each and dissolving them in new microcentrifuge tubes in 174.4 and 270.3 µL of 10 mM pH 7.4 MOPS buffer, respectively, for the reduction and alkylation steps.
- Reduce disulfide bonds by adding 2 µL of the 100 mM TCEP (final concentration of 2 mM in reaction mixture) solution to the reaction mixture and reacting for 3 minutes at room temperature.
NOTE: The final concentration of TCEP should equal 40x the protein concentration per disulfide bond present in the solution. - Alkylate reduced cysteines with 4 µL of the 100 mM IAM solution (final concentration of 4 mM in reaction mixture) for 30 minutes in the dark. IAM is light-sensitive and will decompose under direct light.
NOTE: The final concentration of IAM in solution should be twice the concentration used for TCEP, or 80x the protein concentration per disulfide bond. - Digest the protein with an appropriate enzyme such as trypsin or chymotrypsin. A 10:1 protein:enzyme ratio for a 3-hour digestion at 37 °C with immobilized enzyme at a shaking rate of 300 strokes/min is typically sufficient for DEPC-labeled proteins. See Discussion.
- After digestion, separate the immobilized enzyme from the digested peptides by centrifugation at 12,000 rpm for 5 minutes.
- Analyze the sample immediately by LC-MS/MS or flash-freeze the sample with liquid nitrogen to minimize sample degradation and label loss. Store the flash-frozen samples at < -20 °C until ready for LC-MS/MS analysis.
4. LC-MS/MS Analysis
NOTE: Standard LC-MS/MS parameters for bottom-up proteomics can be used to identify labeled sites on the proteolytic peptide fragments. A general example is outlined below.
- Separate the DEPC-labeled peptides using a reversed-phase C18 stationary phase. Use a typical LC mobile phase of two solvents: (A) water + 0.1% formic acid and (B) ACN + 0.1% formic acid using a gradient (e.g., Figure 2) to achieve the best separation of peptides.
NOTE: The separation time can be optimized based on sample complexity, and mobile phase flow rate depends on whether capillary or nano LC is used. - Use a mass spectrometer capable of doing on-line LC-MS and MS/MS to identify DEPC modification sites on the peptide. In our experiments, we have successfully used several types of mass spectrometers. Any mass spectrometer capable of automatically performing MS/MS of many peptides during the course of an LC-MS analysis should be suitable. Relevant MS parameters include: ESI source voltage = -4000 V for regular ESI; -2000 V for nanospray; Orbitrap resolution = 60,000; Dynamic exclusion duration = 30 s; MS/MS activation type: CID, ETD, or both; Mass scan range = 200-2,000; Automatic Gain Control = 4.0E5 (MS1 in Orbitrap) and 5.0E4 (MS2 in linear quadrupole ion trap).
- Load and inject the digested, labeled protein sample into the LC system and start the LC-MS/MS acquisition. If the sample has been flash-frozen, thaw before analysis. Divert the LC effluent to waste for the first 5 minutes to avoid excessive salts from getting into the ESI source.
NOTE: A 5 µL injection loop is generally utilized, allowing for injection of approximately 2.5 µg of protein to the LC-MS/MS. This is dependent on the loading conditions of the LC as to not clog the sample injector.
5. Data analysis
- Identify DEPC label sites and quantify peptide peak areas using appropriate software for the mass spectrometer that is used.
- Include DEPC addition (72.02 Da) and carbamidomethylation (57.02 Da) as variable modifications. Additional search parameters for the MS/MS analysis are as follows: Maximum missed cleavages = 3; Fragment ion types = b and y; Precursor m/z tolerance = 10 ppm (this value should be higher if a quadrupole ion trap mass spectrometer is used); Fragment m/z tolerance = 0.5 Da (this value should be lower if a high-resolution mass spectrometer is used for a product ion scan); Precursor charge = 1-4.
NOTE: Different database search algorithms have different scoring systems, and many can have difficulty identifying DEPC-modified peptides because modification levels can be low. Adjusting the score cutoff may be necessary to identify more labeled peptides. If so, then manual interrogation of the MS/MS data should be used to verify low-scoring peptides. The product of the hydrolysis of the DEPC label is not included in the data searching because the hydrolyzed DEPC is no longer reactive toward nucleophilic side chains. - Determine residue-level modification percentages using the chromatographic peak areas of the modified and unmodified versions of the peptides.
NOTE: Any peptide containing the modified residue of interest must be considered and all charge states that are included must be present in all the measured samples. Peptides having different ionization efficiencies and eluting at different times causes this value to be a relative rather than absolute measure of the modification of a specific site.
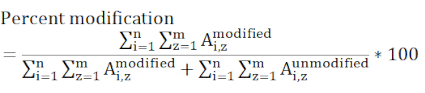
where Ai,z represents peak area of any given peptide (i) that contains the residue of interest and considers all detected charge states (z). - Determine if a labeling change between a control and experimental sample is significant using statistical evaluation. Three replicate measurements for each sample is typical, and t-tests are most commonly utilized with 95 or 99% confidence intervals.
Results
Identifying DEPC modification sites and modification percentages
Mass addition due to covalent labeling can be measured at the (a) intact protein and (b) peptide levels8,9. At the intact level, a distribution of protein species with different numbers of labels can be obtained from direct analysis or LC-MS of labeled protein samples. To obtain higher resolution structural information (i.e., site-specific labeling data), measurements must be performe...
Discussion
Critical Steps
Several points regarding experimental design should be considered to ensure reliable labeling results. First, to maximize protein labeling, it is necessary to avoid buffers with strongly nucleophilic groups (e.g., Tris) because they can react with DEPC and lower the extent of labeling. It is also conceivable that such buffers could react with labeled residues, causing the removal of the label and therefore loss of structural information. We recommend MOPS as a buffer, but phosphate buffered ...
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge support from the National Institutes of Health (NIH) under Grant R01 GM075092. The Thermo Orbitrap Fusion mass spectrometer used to acquire some of the data described here was acquired with funds from the National Institutes of Health grant S10OD010645.
Materials
| Name | Company | Catalog Number | Comments |
| 1.5 mL microcentrifuge tube | Thermo Fisher Scientific | 3448 | |
| 3-(N-morpholino)propanesulfonic acid | Millipore Sigma | M1254 | |
| 3-(N-morpholino)propanesulfonic acid sodium salt | Millipore Sigma | M9381 | |
| Acclaim PepMap RSLC C18 Column | Thermo Scientific | 164537 | 300 μm x 15 cm, C18, 2 μm, 100 A |
| Acetonitrile | Fisher Scientific | A998-1 | |
| Diethylpyrocarbonate | Millipore Sigma | D5758 | |
| HPLC-grade water | Fisher Scientific | W5-1 | |
| Imidazole | Millipore Sigma | I5513 | |
| Immobilized chymotrypsin | ProteoChem | g4105 | |
| Immobilized trypsin, TPCK Treated | Thermo Fisher Scientific | 20230 | |
| Iodoacetamide | Millipore Sigma | I1149 | |
| Tris(2-carboxyethyl)phosphine | Millipore Sigma | C4706 |
References
- Katta, V., Chait, B. T., Carr, S. Conformational Changes in Proteins Probed by Hydrogen-exchange Electrospray-ionization. Rapid Communications in Mass Spectrometry. 5, 214-217 (1991).
- Wales, T. E., Engen, J. R. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrometry Reviews. 25, 158-170 (2006).
- Pirrone, G. F., Iacob, R. E., Engen, J. R. Applications of hydrogen/deuterium exchange MS from 2012 to 2014. Analytical Chemistry. 87, 99-118 (2015).
- Oganesyan, I., Lento, C., Wilson, D. J. Contemporary hydrogen deuterium exchange mass spectrometry. Methods. 144, 27-42 (2018).
- Sinz, A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrometry Reviews. 25, 663-682 (2006).
- Holding, A. N. XL-MS: Protein cross-linking coupled with mass spectrometry. Methods. 89, 54-63 (2015).
- Xu, G., Chance, M. R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chemical Reviews. 107, 3514-3543 (2007).
- Mendoza, V. L., Vachet, R. W. Probing Protein Structure by Amino Acid-Specific Covalent Labeling and Mass Spectrometry. Mass Spectrometry Reviews. 28, 785-815 (2009).
- Limpikirati, P., Liu, T., Vachet, R. W. Covalent labeling-mass spectrometry with non-specific reagents for studying protein structure and interactions. Methods. 144, 79-93 (2018).
- Liu, X. R., Zhang, M. M., Gross, M. L. Mass Spectrometry-Based Protein Footprinting for Higher-Order Structure Analysis: Fundamentals and Applications. Chemistry Reviews. 120, 4335 (2020).
- Maleknia, S. D., Brenowitz, M., Chance, M. R. Millisecond radiolytic modification of peptides by synchrotron X-rays identified by mass spectrometry. Analytical Chemistry. 71, 3965-3973 (1999).
- Aye, T. T., Low, T. Y., Sze, S. K. Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Analytical Chemistry. 77, 5814-5822 (2005).
- Hambly, D. M., Gross, M. L. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. Journal of the American Society of Mass Spectrometry. 16, 2057-2063 (2005).
- Mendoza, V. L., Vachet, R. W. Protein surface mapping using diethylpyrocarbonate with mass spectrometric detection. Analytical Chemistry. 80, 2895-2904 (2008).
- Mendoza, V. L., Antwi, K., Barón-rodríguez, M. A., Blanco, C., Vachet, R. W. Structure of the Pre-amyloid Dimer of β-2-microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 49, 1522-1532 (2010).
- Mendoza, V. L., Barón-Rodríguez, M. A., Blanco, C., Vachet, R. W. Structural insights into the pre-amyloid tetramer of β-2-microglobulin from covalent labeling and mass spectrometry. Biochemistry. 50, 6711-6722 (2011).
- Zhou, Y., Vachet, R. W. Increased protein structural resolution from diethylpyrocarbonate-based covalent labeling and mass spectrometric detection. Journal of the American Society of Mass Spectrometry. 23, 708-717 (2012).
- Borotto, N. B., et al. Investigating Therapeutic Protein Structure with Diethylpyrocarbonate Labeling and Mass Spectrometry. Analytical Chemistry. 87, 10627-10634 (2015).
- Liu, T., Marcinko, T. M., Kiefer, P. A., Vachet, R. W. Using Covalent Labeling and Mass Spectrometry To Study Protein Binding Sites of Amyloid Inhibiting Molecules. Analytical Chemistry. 89, 11583-11591 (2017).
- Limpikirati, P., et al. Covalent labeling and mass spectrometry reveal subtle higher order structural changes for antibody therapeutics. MAbs. 11, 463-476 (2019).
- Limpikirati, P., Pan, X., Vachet, R. W. Covalent Labeling with Diethylpyrocarbonate: Sensitive to the Residue Microenvironment, Providing Improved Analysis of Protein Higher Order Structure by Mass Spectrometry. Analytical Chemistry. 91, 8516-8523 (2019).
- Liu, T., Limpikirati, P., Vachet, R. W. Synergistic Structural Information from Covalent Labeling and Hydrogen-Deuterium Exchange Mass Spectrometry for Protein-Ligand Interactions. Analytical Chemistry. 91, 15248-15254 (2019).
- Pan, X., Limpikirati, P., Chen, H., Liu, T., Vachet, R. W. Higher-Order Structure Influences the Kinetics of Diethylpyrocarbonate Covalent Labeling of Proteins. Journal of the American Society of Mass Spectrometry. 31, 658-665 (2020).
- Limpikirati, P. K., Zhao, B., Pan, X., Eyles, S. J., Vachet, R. W. Covalent Labeling/Mass Spectrometry of Monoclonal Antibodies with Diethylpyrocarbonate: Reaction Kinetics for Ensuring Protein Structural Integrity. Journal of the American Society of Mass Spectrometry. 31, 1223-1232 (2020).
- Liu, T., Marcinko, T. M., Vachet, R. W. Protein-Ligand Affinity Determinations Using Covalent Labeling-Mass Spectrometry. Journal of the American Society of Mass Spectrometry. 31, 1544-1553 (2020).
- Srikanth, R., Mendoza, V. L., Bridgewater, J. D., Zhang, G., Vachet, R. W. Copper Binding to β-2-Microglobulin and its Pre-Amyloid Oligomers. Biochemistry. 48, 9871-9881 (2009).
- Lim, J., Vachet, R. W. Using mass spectrometry to study copper-protein binding under native and non-native conditions: β-2-microglobulin. Analytical Chemistry. 76, 3498-3504 (2004).
- Lindsley, C. W. Predictions and Statistics for the Best-Selling Drugs Globally and in the United States in 2018 and a Look Forward to 2024 Projections. ACS Chemical Neuroscience. 10, 1115 (2019).
- Floege, J., Ketteler, M. β2-Microglobulin-derived amyloidosis: An update. Kidney International. 59, 164 (2001).
- Antwi, K., et al. Cu (II) organizes β-2-microglobulin oligomers but is released upon amyloid formation. Protein Science. 17, 748-759 (2008).
- Dong, J., et al. Unique Effect of Cu(II) in the Metal-Induced Amyloid Formation of β-2-Microglobulin. Biochemistry. 53, 1263-1274 (2014).
- Marcinko, T. M., Drews, T., Liu, T., Vachet, R. W. Epigallocatechin-3-gallate Inhibits Cu(II)-Induced β-2-Microglobulin Amyloid Formation by Binding to the Edge of Its β-Sheets. Biochemistry. 59, 1093-1103 (2020).
- Zhou, Y., Vachet, R. W. Diethylpyrocarbonate Labeling for the Structural Analysis of Proteins: Label Scrambling in Solution and How to Avoid it. Journal of the American Society of Mass Spectrometry. 23, 899-907 (2012).
- Borotto, N. B., Degraan-Weber, N., Zhou, Y., Vachet, R. W. Label scrambling during CID of covalently labeled peptide ions. Journal of the American Society of Mass Spectrometry. 25, 1739-1746 (2014).
- Aprahamian, M. L., Chea, E. E., Jones, L. M., Lindert, S. Rosetta Protein Structure Prediction from Hydroxyl Radical Protein Footprinting Mass Spectrometry Data. Analytical Chemistry. 90, 7721-7729 (2018).
- Schmidt, C., et al. Surface Accessibility and Dynamics of Macromolecular Assemblies Probed by Covalent Labeling Mass Spectrometry and Integrative Modeling. Analytical Chemistry. 89, 1459-1468 (2017).
- Zheng, X., Wintrode, P. L., Chance, M. R. Complementary Structural Mass Spectrometry Techniques Reveal Local Dynamics in Functionally Important Regions of a Metastable Serpin. Structure. 16, 38-51 (2008).
- Pan, Y., Piyadasa, H., O'Neil, J. D., Konermann, L. Conformational dynamics of a membrane transport protein probed by H/D exchange and covalent labeling: The glycerol facilitator. Journal of Molecular Biology. 416, 400-413 (2012).
- Li, J., et al. Mapping the Energetic Epitope of an Antibody/Interleukin-23 Interaction with Hydrogen/Deuterium Exchange, Fast Photochemical Oxidation of Proteins Mass Spectrometry, and Alanine Shave Mutagenesis. Analytical Chemistry. 89, 2250-2258 (2017).
- Borotto, N. B., Zhang, Z., Dong, J., Burant, B., Vachet, R. W. Increased β-Sheet Dynamics and D-E Loop Repositioning Are Necessary for Cu(II)-Induced Amyloid Formation by β-2-Microglobulin. Biochemistry. 56, 1095-1104 (2017).
- Shi, L., Liu, T., Gross, M. L., Huang, Y. Recognition of Human IgG1 by Fcγ Receptors: Structural Insights from Hydrogen-Deuterium Exchange and Fast Photochemical Oxidation of Proteins Coupled with Mass Spectrometry. Biochemistry. 58, 1074-1080 (2019).
- Gerega, S. K., Downard, K. M. PROXIMO - A new docking algorithm to model protein complexes using data from radical probe mass spectrometry (RP-MS). Bioinformatics. 22, 1702-1709 (2006).
- Kamal, J. K. A., Chance, M. R. Modeling of protein binary complexes using structural mass spectrometry data. Protein Science. 17, 79-94 (2007).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved