A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
A Step-By-Step Method to Detect Neutralizing Antibodies Against AAV using a Colorimetric Cell-Based Assay
In This Article
Summary
A comprehensive laboratory protocol and analysis workflow are described for a rapid, cost-effective, and straightforward colorimetric cell-based assay to detect neutralizing elements against AAV6.
Abstract
Recombinant adeno-associated viruses (rAAV) have proven to be a safe and successful vector for transferring genetic material to treat various health conditions in both the laboratory and the clinic. However, pre-existing neutralizing antibodies (NAbs) against AAV capsids pose an ongoing challenge for the successful administration of gene therapies in both large animal experimental models and human populations. Preliminary screening for host immunity against AAV is necessary to ensure the efficacy of AAV-based gene therapies as both a research tool and as a clinically viable therapeutic agent. This protocol describes a colorimetric in vitro assay to detect neutralizing factors against AAV serotype 6 (AAV6). The assay utilizes the reaction between an AAV encoding an alkaline phosphatase (AP) reporter gene and its substrate NBT/BCIP, which generates an insoluble quantifiable purple stain upon combination.
In this protocol, serum samples are combined with an AAV expressing AP and incubated to permit potential neutralizing activity to occur. Virus serum mixture is subsequently added to cells to allow for viral transduction of any AAVs that have not been neutralized. The NBT/BCIP substrate is added and undergoes a chromogenic reaction, corresponding to viral transduction and neutralizing activity. The proportion of area colored is quantitated using a free software tool to generate neutralizing titers. This assay displays a strong positive correlation between coloration and viral concentration. Assessment of serum samples from sheep before and after administration of a recombinant AAV6 led to a dramatic increase in neutralizing activity (125 to >10,000-fold increase). The assay displayed adequate sensitivity to detect neutralizing activity in >1:32,000 serum dilutions. This assay provides a simple, rapid, and cost-effective method to detect NAbs against AAVs.
Introduction
Adeno-associated viruses (AAV) are increasingly used as vectors for the delivery of gene therapies to trial treatments for various health conditions that impact the cardiovascular, pulmonary, circulatory, ocular, and central nervous systems1,2,3,4,5. The popularity of AAV vectors as a leading gene therapy platform stems from their positive safety profile, long-term transgene expression, and wide-ranging tissue-specific tropisms1,6. Successful outcomes in animal studies have paved the way for over fifty AAV gene therapy clinical trials that have successfully reached their efficacy endpoints7, as well as the release of the first commercially available AAV gene therapy drug approved by the US Food and Drug Administration8. Following initial successes, AAV has continued to gain traction in the basic and clinical research sectors as a vector of choice and is currently the only in vivo gene therapy approved for clinical use in the US and Europe9. Nonetheless, the presence of pre-existing neutralizing antibodies (NAbs) against AAV vector capsids remains a hindrance to both preclinical research and the efficacy of clinical trials. NAbs are present in both naïve human and animal populations and inhibit gene transduction following in vivo administration of an AAV vector1. AAV seropositivity is an exclusion criterion for most gene therapy trials, and therefore preliminary screening for host immunity is crucial in both the laboratory and the clinic. Establishing an assay that can detect the presence of NAbs against AAV is an essential step in the pipeline of any AAV gene therapy-based research project. This report focuses on AAV6 which has been of interest to researchers due to its efficient and selective transduction in striated muscle (heart and skeletal muscle)1,10,11,12. Gene therapy is considered a promising strategy for targeting the heart because it is difficult to specifically target the heart without invasive open-heart procedures.
Neutralizing activity is usually determined using either a cell-based in vitro or in vivo transduction inhibition assay. In vivo NAb assays usually involve administering serum from a test subject (e.g., human or large animal) into mice, followed by an AAV with a reporter gene, followed by testing for the expression of the reporter gene or corresponding antigen. In vitro assays determine NAb titers by incubating serum or plasma from a human or large animal in serial dilutions with a recombinant AAV (rAAV) that expresses a reporter gene. Cells are infected with the serum/virus mixture, and the extent to which the reporter gene expression is inhibited is assessed compared with controls. In vitro assays are widely used for NAb screening due to their comparatively lower cost, rapidity in testing, and greater capacity for standardization and validation13,14 compared with in vivo assays. In vivo assays are often reported to have greater sensitivity15,16, but the same claim has been made concerning in vitro assays14,17.
To date, in vitro NAb assays have mainly used luminescence (luciferase) as the reporter gene to detect neutralization. Although a light-based method has merit in many contexts, a colorimetric/chromogenic NAb assay may be advantageous in some circumstances. Colorimetric assays to assess neutralization have been successfully employed for other viruses such as influenza and adenovirus18,19. Their attractiveness stems from their simplicity, lower cost, and the requirement for only everyday laboratory apparatus and tools20. NAb assays that use a luminescence-based reporter gene require costly substrate kits, a luminometer, and corresponding software for analysis21. This colorimetric assay has the advantage of only requiring a light microscope and a very cheap substrate. Reporting of the sensitivity of colorimetric versus luminescent assays has yielded conflicting results. One study suggested luminescence-based ELISA assays display greater sensitivity and comparable reproducibility to colorimetric assays22, while another found colorimetric-based ELISA assays to confer greater sensitivity23. Here, a detailed protocol for an in vitro NAb assay against AAV that utilizes the chromogenic reaction between an AAV encoding an alkaline phosphatase (AP) reporter gene and a nitro blue tetrazolium /5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) substrate is provided. This step-by-step protocol was developed based on a previous report that utilized an hPLAP (human placental alkaline phosphatase) reporter gene (AAV6-hPLAP) to detect neutralizing activity against AAV24. This assay is cost-effective, time-efficient, easy to set up, and requires minimal technical skills, laboratory equipment, and reagents. Moreover, the simplicity of this assay gives it the potential to be optimized for broad applications across different types of cells, tissues, or viral serotypes.
Protocol
All aspects of animal care and experimentation were conducted following Florey Institute of Neuroscience and Mental Health guidelines and the Australian Code for the Care and Use of Animals for Scientific Purposes following Reference25. 1.5-3-year-old Merino ewes were used for the study. A schematic overview of the assay protocol is provided in Figure 1.
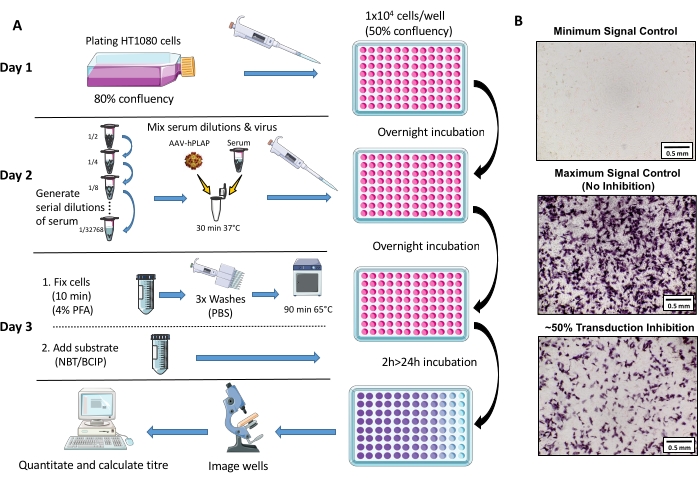
Figure 1: Schematic diagram of NAb assay protocol. (A) Visual representation of the NAb assay illustrating the primary steps involved in the three-day protocol. Briefly, cells are grown and plated overnight. The following day, serial dilutions of serum are prepared, incubated with AAV, and then incubated with the cells overnight. The next day, cells are fixed, washed, incubated, combined with the substrate, and incubated again, followed by imaging and quantitation. (B) Representative images of a minimum signal control (complete AAV inhibition), a maximum signal control (no inhibition), and an ovine serum sample with ~50% signal inhibition. Scale bar = 0.5 mm. Please click here to view a larger version of this figure.
1. Initial preparation
- For assessment in sheep: collect blood in 8 mL serum separator clot activator tubes (see Table of Materials), leave the blood sample at room temperature (RT) for 20-30 min, and subsequently spin down at 2,100 x g for 15 min. The clear supernatant that forms at the top of the tubes is serum. Aliquot the clear aqueous phase into microcentrifuge tubes and store at -80 °C.
NOTE: The serum at -80 °C remains stable for ~5 years. Blood was collected from the carotid vein using a 16 G needle (tip cut-off) and syringe from conscious animals. - Heat inactivate fetal bovine serum (FBS) by placing it in a water bath at 56 °C for 30 min and swirl intermittently. For precision, place a thermometer in a second bottle containing an equivalent volume of water and add it to the heat bath at the same time as the FBS bottle. Begin timing when the thermometer reaches 56 °C.
- Employ proper aseptic technique and cell culture practice for all subsequent steps performed in the cell culture hood26,27. Spray 70% ethanol on all objects and the hood before use and clean with 1% sodium hypochlorite upon completion.
- Make complete Dulbecco's Modified Eagle Medium (DMEM) by combining high glucose (4.5 g/L) DMEM (89%) with heat-inactivated FBS (10%) and Penicillin Streptomycin (1%). Combine and filter using a sterile vacuum filtration system (0.22 µm pore size, polyethersulfone membrane) (see Table of Materials). Store complete DMEM wrapped in foil at 4 °C.
- Establish HT1080 cells (see Table of Materials) and passage in a 75 cm2 square flask as described in Reference28. Create multiple frozen stocks of cells. Do not use cells after 20 passages as further passaging may influence the assay results.
2. Day 1 - Plating of cells
- Passage HT1080 cells when they reach ~80% confluency.
- Pre-warm complete DMEM (prepared in step 1.4), 0.05% trypsin-EDTA, and 1x phosphate-buffered saline (PBS) to 37 °C in a water bath.Remove the growth medium from passaged cells using an aspiration system.
NOTE: All aspiration in this protocol uses a vacuum system with a tube attached to a sterile 5 mL serological pipette. - Wash the cells in 10 mL of pre-warmed (37 °C) 1x PBS and trypsinize cells for 3-4 min in 4 mL of pre-warmed 0.05% trypsin-EDTA to detach the cells from the flask.
- Inactivate the trypsin by adding 6 mL of pre-warmed complete DMEM and pipette the cells into a 50 mL tube. Calculate the number and concentration of viable cells using a hemocytometer and the trypan blue exclusion method29.
- Dilute the cells to a concentration of 1 x 105 cells/mL in pre-warmed complete DMEM. Seed 100 µL of cells/well into clear 96-well flat-bottomed plates (1 x 104 cells per well). Incubate the plate at 37 °C, 5% carbon dioxide (CO2) overnight for 16-22 h.
3. Day 2 - Infecting the cells
- Remove plate/s from the incubator and use a light microscope to confirm that cells are evenly dispersed within the wells and that the confluency is ~50%. If cells are not within a range of 45%-55% confluency, repeat the 'Day 1' protocol and adjust initial cell concentration accordingly.
- Generate serial dilutions of the serum samples of interest in 1.5 mL microcentrifuge tubes using pre-warmed complete DMEM as the diluent. Table 1 demonstrates the generation of a dilution cascade for triplicate samples.
- To perform the assay in triplicate, prepare a 7.5 x 106 vector genomes (vg)/µL of working solution of AAV6-hPLAP (see Table of Materials) by diluting a virus stock solution in 1x PBS.
- Add 66 µL of the 7.5 x 106 vg/µL virus working solution to each tube containing 264 µL of serum/media dilution (330 µL of total volume/dilution, see Table 1).
NOTE: This is a robust assay that does not require perfect culture conditions. However, to accurately quantitate and ensure each assay run is reliable, it is necessary to include the following: (1) a virus and media only control, (2) a media only control, and (3) a NAb positive control sample on all plates under the same experimental conditions. The volume described (330 µL) accounts for triplicate samples +10% of the serum and virus mixture. Performing replicates is highly recommended for the accurate determination of neutralizing activity.
- Mix the virus/serum dilutions by pipetting and place the tubes containing the virus/serum mixtures in an incubator at 37 °C, 5% CO2 for 30 min to allow potential neutralization to occur.
- Pipette 100 µL of the virus/serum mixture to each well on the 96-well plate containing 1 x 104 cells/well for each dilution.
NOTE: This will generate a final viral concentration of 15k viruses/cell multiplicity of infection (MOI) in each well. Table 2 provides an example 96-well sample plate layout for assessing samples to a 1/512 dilution. - Wrap the 96-well plate containing cells, serum, and AAV-hPLAP in foil and place in an incubator at 37 °C, 5% CO2 overnight for 16-24 h to allow AAV entry into the cells.
| Dilution cascade label | Dilution | 3 x sample (240 μL) + 10% buffer volume (24 μL) | Ratio of serum:media |
| Dilution 1 (D1) | 1/2 | 264 μL serum 264 μL media | 50:50 |
| Dilution 2 (D2) | 1/4 | 264 μL D1 + 264 μL media | 25:75 |
| Dilution 3 (D3) | 1/8 | 264 μL D2 +264μL media | 12.5:87.5 |
| Dilution 4 (D4) | 1/16 | 264 μL D3 +264 μL media | 6.25:93.75 |
| Dilution 5 (D5) | 1/32 | 264 μL D4 +264 μL media | 3.13:96.87 |
| Dilution 6 (D6) | 1/64 | 264 μL D5 +264 μL media | 1.56:98.44 |
| Dilution 7 (D7) | 1/128 | 264 μL D5 +264 μL media | 0.78:99.22 |
| Dilution 8 (D8) | 1/256 | 264 μL D5 +264 μL media | 0.39:99.61 |
| Dilution 9 (D9) | 1/512 | 264 μL D7 + 264 μL media | 0.2:99.8 |
| Dilution 10 (D10) | 1/2048 | 132 μL D8 + 396 μL media | 0.05:99.95 |
| Dilution 11 (D11) | 1/8192 | 132 μL D9 + 396 μL media | 0.01:99.99 |
| Dilution 12 (D12) | 1/32768 | 132 μL D10 + 396 μL media | 0.003:99.997 |
Table 1: Volumes of serum and diluent required to generate serial dilutions of serum in triplicate.
| Serum sample #1 | Serum sample #2 | Serum sample #3 | Mono AB (mAB), controls and extra samples | |||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 50 ng MAb | 50 ng MAb | 50 ng MAb |
| B | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 5 ng MAb | 5 ng MAb | 5 ng MAb |
| C | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 0.5 ng MAb | 0.5 ng MAb | 0.5 ng MAb |
| D | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | MO (-C) | MO (-C) | MO (-C) |
| E | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | VO (+C) | VO (+C) | VO (+C) |
| F | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | Sample #1 1/512 | Sample #1 1/512 | Sample #1 1/512 |
| G | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | Sample #2 1/512 | Sample #2 1/512 | Sample #2 1/512 |
| H | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | Sample #3 1/512 | Sample #3 1/512 | Sample #3 1/512 |
Table 2: Example 96-well plate layout for assessing naïve serum samples in dilutions ranging from 1/2 to 1/512. Higher dilutions are incorporated into the assay if assessing a sample known to be positive for AAV NAbs (post-administration samples) or if a higher titer is required. MO (-C): Media-only control. VO (+C): Virus and media only control. mAb: Monoclonal antibody against AAV (NAb positive control).
4. Day 3 - Fixing and adding substrate to the cells
- Pre-warm an aliquot of 1x PBS to 37 °C (~25 mL/96-well plate). Cool separate aliquots of PBS (~25 mL/96-well plate) and double-distilled H2O (DDW, ~25 mL/96-well plate) to 4 °C. Dissolve a pellet of BCIP/NBT (see Table of Materials) in 10 mL of DDW in a 50 mL conical centrifuge tube by vortexing (10 mL is enough for 2 x 96 well plates).
- Aspirate the media from the wells of the 96-well plate using a serological pipette or similar attached to a suction-based aspiration system or fume hood vacuum. Gently place the tip of the serological pipette into the well and remove the media taking caution not to disrupt the adhered cells.
- Add 50 µL of RT 4% PFA to each well using a pipette. Wrap the plate in foil and leave it at RT for 10 min to fix the cells.
CAUTION: Paraformaldehyde (PFA) is a probable carcinogen and is toxic from skin or eye contact or inhalation. Handle in a fume hood with proper personal protective equipment as well as a facemask. Make fresh 4% PFA diluted in PBS (~7 mL required per 96-well plate).
- Add 50 µL of RT 4% PFA to each well using a pipette. Wrap the plate in foil and leave it at RT for 10 min to fix the cells.
- Wash and aspirate the cells with 200 µL of RT 1x PBS. Repeat this step twice.
NOTE: A multichannel pipette is an efficient option for the pipetting steps. - Pipette 200 µL of pre-warmed PBS into each well, wrap the plate in foil and incubate at 65 °C for 90 min to denature endogenous alkaline phosphatase activity30.
- Aspirate wells and wash cells with 200 µL of cold (4 °C) PBS. Aspirate again, wash in 200 µl of cold DDW, and aspirate again.
- Pipette 50 µL of the dissolved BCIP/NBT (prepared in step 4.1) into each well.
- Wrap the plate in foil and incubate at RT for 2-24 h.
NOTE: Be consistent with incubation time between runs; the time flexibility allows users to photograph wells either on day 3 or the following day. - Using a light microscope camera, take photos of each well using a 4x objective lens, ensuring the same exposure, white balancing, and light settings are used consistently for all assays performed.
- Position each well identically and ensure the edges of the well are not visible in the photos. Save photos in TIF format or similar.
NOTE: Specific settings will vary between microscopes, but quantitation will be most effective if the background lighting is high and consistent throughout the wells (Figure 1B).
- Position each well identically and ensure the edges of the well are not visible in the photos. Save photos in TIF format or similar.
5. Quantitation to determine the neutralizing activity using ImageJ
- Download and install the freely available software "ImageJ" (see Table of Materials).
- Open the image to be analyzed in ImageJ by selecting File > Open (Figure 2).
- If using colored images, convert to grayscale by selecting Image > Type > 8-bit.
- Click on Image > Adjust > Threshold. Adjust the threshold until all colored areas are colored in red, but the background is not. Upon adding NBT/BCIP, the colored product will deposit in the area around the cells expressing hPLAP.
NOTE: It is recommended to use the same threshold setting for all images captured on the same plate. - Click on Analyze > Set Measurements and tick the Area, Limit to Threshold, Area Fraction, and Display label checkboxes and click on Ok.
- To determine the signal reading (percentage of coloration) of a given well, click on Analyze > Measure. The '% Area' column of the pop-up window displays the signal reading.
- Perform quantitation for all sample replicates. Exclude any contaminated wells, wells showing uneven cell distribution, or wells varying in cell density or lighting.
NOTE: See Supplementary Figure 1 for examples of wells that should be considered for exclusion. Typically, 3-4 wells may require exclusion from a 96-well plate. Figure 2 provides a visual representation of the quantitation process using ImageJ.
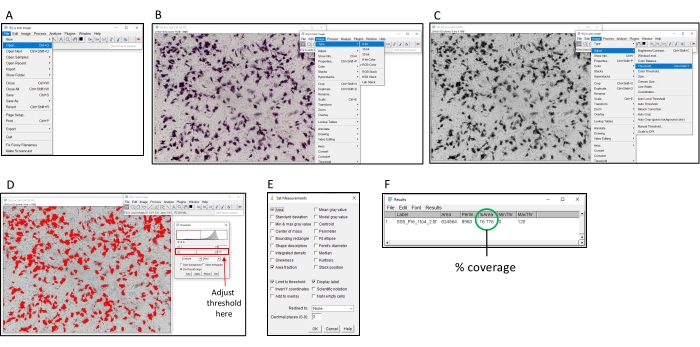
Figure 2: Steps for determining percentage coloration using ImageJ software. (A) Open the image to be analyzed with ImageJ software. (B) Convert the image to 8-bit grayscale. (C) Open the threshold window. (D) Adjust the maximum threshold so all colored areas are covered, but the background area is not (this threshold should be consistent across an entire plate). (E) Select the 'Analyze' dropbox, click on 'Set measurements' and tick 'Area', 'Area fraction', 'Limit threshold' and 'Display label', and click on 'OK'. (F) Click on 'Measure' to measure the covered area. The % area indicates the proportion of the image that was colored. This can then be used with the control samples to determine the TI50 titer. Please click here to view a larger version of this figure.
6. Determination of Transduction Inhibition (TI50) titer
- Determine the average readout from replicates (using the steps described in step 5) for the following: (1) Media-only control (baseline signal reading). (2) Virus + media only control (maximum signal reading). (3) Virus + serum samples of interest.
- Calculate the percentage of inhibition using the following formula:
100 - [(Test sample signal readout (virus + serum sample of interest) - baseline signal readout (media only control)) / (maximum signal readout (media and virus only) - baseline signal readout) x 100] = % Transduction inhibition13. - Calculate the % transduction inhibition from all replicates of each dilution for all samples using the formula in 6.2. Determine the average transduction inhibition between the technical replicates for each dilution for all samples and controls.
- Calculate the 50% transduction inhibition titer (TI50 titer) of a sample of interest by determining the lowest dilution of the sample that yields 50% or greater transduction inhibition of hPLAP activity. e.g., if a 1/8 dilution of a sample has greater than 50% transduction inhibition based on the calculation performed in 6.2 (and a 1/4 dilution does not), report the TI50 titer as 1/8.
7. Determination of neutralized AAV particles
- Calculate the number of neutralized AAV particles per µL of serum for a given sample by employing the following formula:
((MOI x cell count/well) / (volume of serum / dilution factor of TI50 titer)) / 2 = neutralized AAV particles / µL of serum9.
NOTE: Dividing by 2 accounts for the TI50 measuring 50% of neutralized particles. For a sample that gives a TI50 titer of 1/4 (25% serum, 75% diluent) in which the assay used 80 μL of undiluted serum and an MOI of 15k plated onto 1 x 104 cells, the following calculation would be used: ((15000 x 10000) / (80/4)) / 2 = 3.75x106 neutralized particles / µL of serum.
Results
Transduction assay to establish the optimal viral dosage for plate coverage
HT1080 cells, a well-established fibrosarcoma cell line, were selected for this assay. A concentration of 1 x 104 HT1080 cells/well provided ~50% cell confluency in each well of a 96-well plate. To determine the optimal viral concentration for the assay, an rAAV encoding an hPLAP (human placental alkaline phosphatase) reporter gene (AAV6-hPLAP)31 was added in triplicate at a range of concent...
Discussion
This report describes a colorimetric assay that assesses the extent of AAV neutralization in a given serum sample by evaluating a chromogenic reaction corresponding to the degree of in vitro viral transduction. The development of the protocol was based on the known chromogenic reaction between the enzyme alkaline phosphatase and NBT/BCIP, which has been widely utilized as a staining tool for the detection of protein targets in applications such as immunohistochemistry and as a reporter tool for evaluating viral ...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was funded by a National Health and Medical Research Council Project Grant to JRM and CJT (ID 1163732) and in part by the Victorian Government's Operational Infrastructure Support Program. SB is supported by a joint Baker Heart and Diabetes Institute-La Trobe University Doctoral Scholarship. KLW is supported by The Shine On Foundation and a Future Leader Fellowship from the National Heart Foundation of Australia (ID 102539). JRM is supported by a National Health and Medical Research Council Senior Research Fellowship (ID 1078985).
Materials
| Name | Company | Catalog Number | Comments |
| 0.05% Trypsin/EDTA | Gibco | 25300-054 | |
| 50 mL conical centrifuge tube | Falcon | 14-432-22 | Or equivalent |
| 75 cm2 square flasks | Falcon | 353136 | Or equivalent |
| 96 well flat bottomed plate | Falcon | 353072 | |
| AAV6-CMV-hPLAP Vector | Muscle Research & Therapeutics Lab (University of Melbourne, Australia) AAV6-CMV-hPLAP can be provided upon request. | ||
| Aluminium foil | |||
| Anti-AAV6 (intact particle) mouse monoclonal antibody, (ADK6) | PROGEN | 610159 | Positive control monoclonal antibody |
| BCIP/NBT | SIGMAFAST | B5655 | |
| Cell and tissue culture safety cabinet | |||
| Electronic Pipette | 5 & 10 mL stripette inserts | ||
| Fetal Bovine Serum | Gibco | 10099-141 | |
| Haemocytometer | |||
| High glucose Dulbecco's Modified Eagle Medium (DMEM) | Gibco | 11965118 | |
| HT1080 cells | ATCC | ||
| ImageJ Software | Freely available: https://imagej.nih.gov/ij/download.html | ||
| Incubator | 37 °C, 5% CO2 | ||
| Light microscope with camera | Capable of taking photos with a 4x objective lens | ||
| Oven | For a 65 °C incubation | ||
| Paraformaldehyde | MERCK | 30525-89-4 | |
| Penicillin Streptomycin | Gibco | 15140-122 | |
| Phosphate buffered saline | |||
| Pipettes and tips | 20 μL, 200 μL & 1 mL single pipettes and tips & 200 μL multichannel pipette | ||
| Stericup quick release filter | Millipore | S2GPU10RE | Used for combining media reagents |
| Trypan blue solution | Sigma-Aldrich | T8154 | |
| VACUETTE TUBE 8 ml CAT Serum Separator Clot Activator | Greiner BIO-ONE | 455071 | Used for serum collection & processing from sheep |
| Water bath |
References
- Bass-Stringer, S., et al. Adeno-associated virus gene therapy: Translational progress and future prospects in the treatment of heart failure. Heart, Lung and Circulation. 27 (11), 1285-1300 (2018).
- Casey, G. A., Papp, K. M., MacDonald, I. M. Ocular gene therapy with adeno-associated virus vectors: current outlook for patients and researchers. Journal of Ophthalmic and Vision Research. 15 (3), 396-399 (2020).
- Lykken, E. A., Shyng, C., Edwards, R. J., Rozenberg, A., Gray, S. J. Recent progress and considerations for AAV gene therapies targeting the central nervous system. Journal of Neurodevelopmental Disorders. 10 (1), 16 (2018).
- Guggino, W. B., Cebotaru, L. Adeno-Associated Virus (AAV) gene therapy for cystic fibrosis: Current barriers and recent developments. Expert Opinion on Biological Therapy. 17 (10), 1265-1273 (2017).
- Perrin, G. Q., Herzog, R. W., Markusic, D. M. Update on clinical gene therapy for hemophilia. Blood. 133 (5), 407-414 (2019).
- Wang, D., Tai, P. W. L., Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nature Reviews Drug Discovery. 18 (5), 358-378 (2019).
- Kuzmin, D. A., et al. The clinical landscape for AAV gene therapies. Nature Reviews Drug Discovery. 20 (3), 173-174 (2021).
- Russell, S., et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: A randomised, controlled, open-label, phase 3 trial. Lancet. 390 (10097), 849-860 (2017).
- Weber, T. Anti-AAV Antibodies in AAV gene therapy: Current challenges and possible solutions. Frontiers in Immunology. 12, 658399 (2021).
- Weeks, K. L., et al. Phosphoinositide 3-kinase p110alpha is a master regulator of exercise-induced cardioprotection and PI3K gene therapy rescues cardiac dysfunction. Circulation: Heart Failure. 5 (4), 523-534 (2012).
- Gregorevic, P., et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nature Medicine. 10 (8), 828-834 (2004).
- Bernardo, B. C., et al. Gene delivery of medium chain acyl-coenzyme A dehydrogenase induces physiological cardiac hypertrophy and protects against pathological remodelling. Clinical Science (London). 132 (3), 381-397 (2018).
- Meliani, A., et al. Determination of anti-adeno-associated virus vector neutralizing antibody titer with an in vitro reporter system. Human Gene Therapy Methods. 26 (2), 45-53 (2015).
- Falese, L., et al. Strategy to detect pre-existing immunity to AAV gene therapy. Gene Therapy. 24 (12), 768-778 (2017).
- Wang, D., et al. Adeno-Associated virus neutralizing antibodies in large animals and their impact on brain intraparenchymal gene transfer. Molecular Therapy - Methods & Clinical Development. 11, 65-72 (2018).
- Wang, M., et al. Prediction of adeno-associated virus neutralizing antibody activity for clinical application. Gene Therapy. 22 (12), 984-992 (2015).
- Kruzik, A., et al. Detection of biologically relevant low-titer neutralizing antibodies against adeno-associated virus require sensitive in vitro assays. Human Gene Therapy Methods. 30 (2), 35-43 (2019).
- Lehtoranta, L., Villberg, A., Santanen, R., Ziegler, T. A novel, colorimetric neutralization assay for measuring antibodies to influenza viruses. Journal of Virological Methods. 159 (2), 271-276 (2009).
- Johnston, P. B., Grayston, J. T., Loosli, C. G. Adenovirus neutralizing antibody determination by colorimetric assay. Proceedings of the Society for Experimental Biology and Medicine. 94 (2), 338-343 (1957).
- Xiaoli Zhu, T. G. . Nano-Inspired Biosensors for Protein Assay with Clinical Applications. , 237-264 (2019).
- Jungmann, A., Muller, O., Rapti, K. Cell-based measurement of neutralizing antibodies against adeno-associated virus (AAV). Methods in Molecular Biology. 1521, 109-126 (2017).
- Samineni, S., et al. Optimization, comparison, and application of colorimetric vs. chemiluminescence based indirect sandwich ELISA for measurement of human IL-23. Journal of Immunoassay and Immunochemistry. 27 (2), 183-193 (2006).
- Siddiqui, J., Remick, D. G. Improved sensitivity of colorimetric compared to chemiluminescence ELISAs for cytokine assays. Journal of Immunoassay and Immunochemistry. 24 (3), 273-283 (2003).
- Arnett, A. L., Garikipati, D., Wang, Z., Tapscott, S., Chamberlain, J. S. Immune responses to rAAV6: The Influence of canine parvovirus vaccination and neonatal administration of viral vector. Frontiers in Microbiology. 2, 220 (2011).
- Australian code for the care and use of animals for scientific purposes. National Health and Medical Research Council Available from: https://www.nhmrc.gov.au/about-us/publications/australian-code-care-and-use-animals-scientific-purposes (2013)
- Coecke, S., et al. Guidance on good cell culture practice. A report of the second ECVAM task force on good cell culture practice. Alternatives to Laboratory Animals. 33 (3), 261-287 (2005).
- Journal of Visualized Experiments. General Laboratory Techniques. Journal of Visualized Experiments Database. , (2018).
- AAV-HT1080 Cells. Stratagene Available from: https://www.chem-agilent.com/pdf/strata/240109.pdf (2003)
- Strober, W. Trypan blue exclusion test of cell viability. Current Protocols in Immunology. 111 (3), 1-3 (2015).
- Bieber, S., et al. Extracorporeal delivery of rAAV with metabolic exchange and oxygenation. Scientific Reports. 3, 1538 (2013).
- Winbanks, C. E., Beyer, C., Qian, H., Gregorevic, P. Transduction of skeletal muscles with common reporter genes can promote muscle fiber degeneration and inflammation. PLoS One. 7 (12), 51627 (2012).
- Thomas, C. J., et al. Evidence that the MEK/ERK but not the PI3K/Akt pathway is required for protection from myocardial ischemia-reperfusion injury by 3',4'-dihydroxyflavonol. European Journal of Pharmacology. 758, 53-59 (2015).
- Barger, A., et al. Use of alkaline phosphatase staining to differentiate canine osteosarcoma from other vimentin-positive tumors. Veterinary Pathology. 42 (2), 161-165 (2005).
- Gregorevic, P., et al. Evaluation of vascular delivery methodologies to enhance rAAV6-mediated gene transfer to canine striated musculature. Molecular Therapy. 17 (8), 1427-1433 (2009).
- Sharma, A., Ghosh, A., Hansen, E. T., Newman, J. M., Mohan, R. R. Transduction efficiency of AAV 2/6, 2/8 and 2/9 vectors for delivering genes in human corneal fibroblasts. Brain Research Bulletin. 81 (2-3), 273-278 (2010).
- Smejkal, G. B., Kaul, C. A. Stability of nitroblue tetrazolium-based alkaline phosphatase substrates. Journal of Histochemistry & Cytochemistry. 49 (9), 1189-1190 (2001).
- Falese, L., et al. Strategy to detect pre-existing immunity to AAV gene therapy. Gene Therapy. 24 (12), 768-778 (2017).
- Orlowski, A., et al. Successful transduction with AAV Vectors after selective depletion of anti-aav antibodies by immunoadsorption. Molecular Therapy - Methods & Clinical Development. 16, 192-203 (2020).
- Goossens, K., et al. Differential microRNA expression analysis in blastocysts by whole mount in situ hybridization and reverse transcription quantitative polymerase chain reaction on laser capture microdissection samples. Analytical Biochemistry. 423 (1), 93-101 (2012).
- Entrican, G., Wattegedera, S. R., Griffiths, D. J. Exploiting ovine immunology to improve the relevance of biomedical models. Molecular Immunology. 66 (1), 68-77 (2015).
- Walters, E. M., Prather, R. S. Advancing swine models for human health and diseases. Molecular Medicine. 110 (3), 212-215 (2013).
- Rapti, K., et al. Neutralizing antibodies against AAV serotypes 1, 2, 6, and 9 in sera of commonly used animal models. Molecular Therapy. 20 (1), 73-83 (2012).
- Tellez, J., et al. Characterization of naturally-occurring humoral immunity to AAV in sheep. PLoS One. 8 (9), 75142 (2013).
- Gupta, S., et al. Recommendations for the validation of cell-based assays used for the detection of neutralizing antibody immune responses elicited against biological therapeutics. Journal of Pharmaceutical and Biomedical Analysis. 55 (5), 878-888 (2011).
- Gupta, S., et al. Recommendations for the design, optimization, and qualification of cell-based assays used for the detection of neutralizing antibody responses elicited to biological therapeutics. Journal of Immunological Methods. 321 (1-2), 1-18 (2007).
- Shankar, G., et al. Recommendations for the validation of immunoassays used for detection of host antibodies against biotechnology products. Journal of Pharmaceutical and Biomedical Analysis. 48 (5), 1267-1281 (2008).
- U.S. Department of Health and Human Services Food and Drug Administration. Center for Drug Evaluation and Research (CDER). Immunogenicity Testing of Therapeutic Protein Products — Developing and Validating Assays for Anti-Drug Antibody Detection. U.S. Department of Health and Human Services Food and Drug Administration. , (2019).
- Baatartsogt, N., et al. A sensitive and reproducible cell-based assay via secNanoLuc to detect neutralizing antibody against adeno-associated virus vector capsid. Molecular Therapy - Methods & Clinical Development. 22, 162-171 (2021).
- Watano, R., Ohmori, T., Hishikawa, S., Sakata, A., Mizukami, H. Utility of micro mini pigs for evaluating liver-mediated gene expression in the presence of neutralizing antibody against vector capsid. Gene Therapy. 27 (9), 427-434 (2020).
- Majowicz, A., et al. Therapeutic hFIX activity achieved after single AAV5-hFIX treatment in Hemophilia B patients and NHPs with pre-existing anti-AAV5 NABs. Molecular Therapy - Methods & Clinical Development. 14, 27-36 (2019).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved