A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Evaluating Autophagy Levels in Two Different Pancreatic Cell Models Using LC3 Immunofluorescence
In This Article
Summary
The goal of this protocol is to determine autophagic levels in pancreatic cancer and pancreatic acinar cells through LC3 immunofluorescence and LC3 dot quantification.
Abstract
Autophagy is a specialized catabolic process that selectively degrades cytoplasmic components, including proteins and damaged organelles. Autophagy allows cells to physiologically respond to stress stimuli and, thus, maintain cellular homeostasis. Cancer cells might modulate their autophagy levels to adapt to adverse conditions such as hypoxia, nutrient deficiency, or damage caused by chemotherapy. Ductal pancreatic adenocarcinoma is one of the deadliest types of cancer. Pancreatic cancer cells have high autophagy activity due to the transcriptional upregulation and post-translational activation of autophagy proteins.
Here, the PANC-1 cell line was used as a model of pancreatic human cancer cells, and the AR42J pancreatic acinar cell line was used as a physiological model of highly differentiated mammalian cells. This study used the immunofluorescence of microtubule-associated protein light chain 3 (LC3) as an indicator of the status of autophagy activation. LC3 is an autophagy protein that, in basal conditions, shows a diffuse pattern of distribution in the cytoplasm (known as LC3-I in this condition). Autophagy induction triggers the conjugation of LC3 to phosphatidylethanolamine on the surface of newly formed autophagosomes to form LC3-II, a membrane-bound protein that aids in the formation and expansion of autophagosomes. To quantify the number of labeled autophagic structures, the open-source software FIJI was utilized with the aid of the "3D Objects Counter" tool.
The measure of the autophagic levels both in physiological conditions and in cancer cells allows us to study the modulation of autophagy under diverse conditions such as hypoxia, chemotherapy treatment, or the knockdown of certain proteins.
Introduction
Macroautophagy (commonly referred to as autophagy) is a specialized catabolic process that selectively degrades cytoplasmic components, including proteins and damaged organelles1,2. Autophagy allows cells to physiologically respond to stress stimuli and, thus, maintain cellular homeostasis3. During autophagy, a double membrane vesicle is formed: the autophagosome. The autophagosome contains the cargo molecules and drives them to the lysosome for degradation1,4.
Autophagosomes are decorated by the autophagic protein microtubule-associated protein light chain 3 (LC3)5. When autophagy is not induced, LC3 is diffused in the cytoplasm and nucleus in the LC3-I conformation. On the other hand, when autophagy is induced, LC3 is conjugated with a phosphatidylethanolamine in the membrane of the autophagic structures6. This new LC3 conformation is known as LC3-II1. The LC3 conformation shift causes changes in its cellular localization and its dodecyl sodium sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) migration, which can be detected by techniques such as immunofluorescence and western blot5,7. In this way, LC3 conjugation is a key event in the autophagic process that can be used to measure autophagic activity.
The pancreatic acinar cell is a highly differentiated cell that, under healthy conditions, has a low rate of autophagy. However, in different physiological conditions or under pharmacological stimulation, they can activate autophagy. Therefore, the determination of autophagic levels in this cell line is useful for studying the potential direct or indirect effects of different pharmacological or biological agents on autophagy8,9.
Ductal pancreatic adenocarcinoma is one of the deadliest types of cancer, given its late diagnosis and its high chemotherapy resistance10. Pancreatic cancer cells have high autophagy activity due to the transcriptional upregulation and post-translational activation of autophagy-related proteins11. Pancreatic cancer cells may adjust their autophagy levels in response to unfavorable conditions like hypoxia, nutrient deprivation, or chemotherapy-induced damage11. Hence, analyzing the autophagy levels in pancreatic cancer cells can help understand how they adapt to varying environments and evaluate the effectiveness of autophagy-modulating treatments.
This study shows a method to perform LC3 immunofluorescence in two distinct pancreatic cellular models. The first model, PANC-1 cells, served as a model for pancreatic ductal adenocarcinoma. These cells were treated with gemcitabine, a chemotherapy agent that has previously been shown to induce autophagy, specifically in pancreatic cancer cells carrying the oncogenic Kirsten rat sarcoma virus gene (KRAS)12,13. The second model, AR42J cells, served as a more physiological model of exocrine pancreatic cells. These cells were differentiated with dexamethasone to become more similar to acinar pancreatic cells14. In these cells, autophagy was pharmacologically induced through the use of PP242, which is a potent mTOR inhibitor15. In this study, we demonstrate the applicability of the protocol described with two different pancreatic models and its ability to discriminate between states of low and high autophagy.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Cell preparation
- Soak 12 mm round coverslips in absolute ethanol, and place them vertically in the wells of a 24-well plate.
- Remove the cover, and expose the multi-well plate to ultraviolet radiation for 15 min.
- Position the coverslips horizontally, and wash them with Dulbecco's Modified Eagle Medium (DMEM).
- Seed a low passage number of pancreatic cells. The amount should be adjusted to obtain 50%-75% confluency on the day of fixation16.
NOTE: It is recommended to seed 2.5 × 104 PANC-1 or 4 × 104 AR42J cells per well to fixate the cells after 3 days. - Culture the cells in DMEM containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin in an incubator at 37 °C under a humidified atmosphere with 5% carbon dioxide (CO2).
NOTE: For PANC-1 cells, it is recommended to incubate the cells for 2 days between cell seeding and the following steps. After this time, the cells can be transfected, treated, or fixated. This protocol exemplifies the treatment with gemcitabine in non-transfected PANC-1 cells and the differentiation and PP242 treatment for non-transfected AR42J cells.
2. Treating the cells
- Gemcitabine treatment for PANC-1 cells
- Prepare a solution of 1 µg/µL gemcitabine in DMEM 2 days after seeding. Treat each well with 2.6 µL of the 1 µg/µL gemcitabine solution to achieve a final dilution of 20 µM.
- Incubate the cells for 24 h in the incubator.
- AR42J differentiation and PP242 treatment
- Prepare a solution of 4 µg/mL dexamethasone in DMEM.
- Treat each well with 4.9 µL of 4 µg/mL dexamethasone solution to obtain a final dilution of 100 nM.
- Incubate the cells for 48 h in the incubator.
- Remove the medium, and treat each well with 0.5 µL of 1 mM PP242 to obtain a final dilution of 1 µM.
- Incubate the cells for 2 h in the incubator.
3. Fixing and permeabilizing the cells
- Prepare a 24-well plate with cold methanol and a 6-well plate with cold phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 2 mM KH2PO4). Maintain them on ice.
- Take each coverslip with tweezers, wash it twice in PBS, and incubate for 6 min in methanol.
4. Blocking the cells
- Wash each coverslip twice in PBS, and incubate for 1 h in 10% fetal bovine serum in PBS (blocking solution).
NOTE: In this step, the protocol might be paused. The coverslips can be stored overnight in the fridge in the blocking solution, and the protocol can be continued the following day.
5. Incubating the coverslips with the primary antibody
- Prepare a 1:1,000 solution of anti-LC3 in the blocking solution, and maintain it on ice.
- Place a piece of laboratory sealing film over the multi-well lid.
- Place one drop (25 µL) per coverslip of anti-LC3 solution over the sealing film.
- Take each coverslip with tweezers, and place it over the primary antibody drop, taking care that the cell side is in contact with the solution.
- Prepare a humid chamber by placing a humid piece of paper into a flat-bottom plastic box.
- Place the multi-well plate into the humidity chamber, cover it with foil, and incubate overnight in the fridge.
6. Incubating the coverslips with the secondary antibody
- Remove the multi-well plate from the humidity chamber, and place the coverslips back in the multi-well plate.
- Perform three washes with PBS.
- Prepare a solution of fluorescently labeled anti-rabbit with a dilution of 1:800 in the blocking solution, and maintain it on ice protected from light.
- Place a sealing film piece over the multi-well lid.
- Place a drop (25 µL) per coverslip of anti-rabbit solution over the sealing film.
- Take each coverslip with tweezers, and place it over the primary antibody drop, taking care that the cell side is in contact with the solution.
- Incubate the multi-well plate in the humidity chamber for 2 h at room temperature (RT) protected from light.
7. Staining the cells with 4′ ,6-diamidino-2-phenylindole (DAPI)
- Remove the multi-well plate from the humidity chamber, and place the coverslips back in the multi-well plate.
- Perform three washes with PBS.
- Prepare a 300 nM solution of DAPI in PBS (protected from light).
- Incubate each coverslip with the DAPI solution for 10 min.
- Perform three washes with PBS. Maintain the multi-well plate protected from light.
8. Montage
- Prepare two beakers with water and a piece of paper.
- Place one drop (10 µL) per coverslip of a polyvinyl alcohol-Bis(trimethylaluminum)-1,4-diazabicyclo[2.2.2]octane adduct (PVA-DABCO) solution on a slide.
NOTE: PVA-DABCO is prepared by combining 0.25 M DABCO, 10% W/V PVA, 20% glycerol, and 50% Tris HCl (1.5 M, pH 8.8) in ultrapure water. - Take each coverslip with tweezers, wash it in each water beaker, dry it off in the paper, and place it over the PVA-DABCO drop (with the cells in contact with the solution).
- Let it dry overnight, protected from light.
9. Confocal microscopy viewing and image capture
- Visualize the coverslips in an inverted confocal microscope using an objective of around 63x17.
- Capture representative images of the labeled cells.
10. Quantifying the LC3 dots
- Drag and drop each image file containing the captured channels, such as ".czi", into the ImageJ (FIJI) screen to open. Click on Ok in the dialog box, and close the Console window.
- From the Image tab, select Color > Split Channels.
- Close the images corresponding to the channels other than the LC3 image.
- From the Image tab, select Adjust > Color Balance
- Move the Maximum slider to the left until the image is saturated to visualize the cell contours.
- Draw the cell outline with the Freehand Selection tool.
- Click on the Reset button to reset the color adjustment.
- From the Edit tab, select Cut to cut the selected item.
- Close the image without saving it.
- From the Edit tab, select Paste.
- In the Analyze menu, choose the tool 3D Objects Counter.
- Set the threshold. In the example provided in this study, the threshold is set at 2,000.
- Set the size filter. In this study, it is set between 50 and 500.
- Be sure that the boxes Objects and Summary are marked.
- Click on Ok. The number of dots will be described as Objects Detected in the Summary.
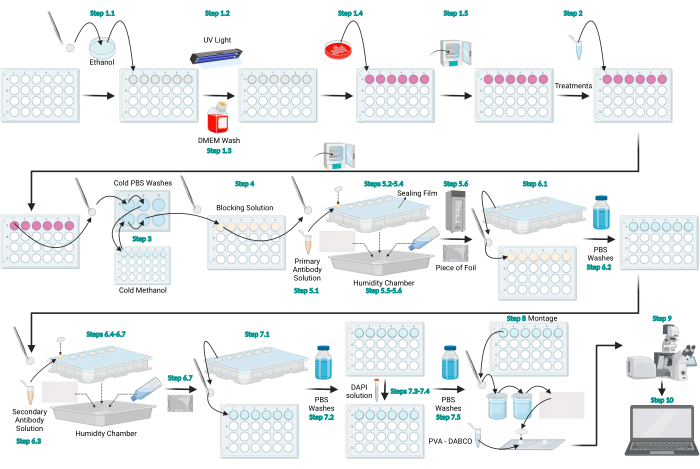
Figure 1: Schematic diagram of the LC3 immunofluorescence protocol. Schematic diagram that represents the general protocol provided for LC3 immunofluorescence. Figure created with BioRender.com. Please click here to view a larger version of this figure.
Access restricted. Please log in or start a trial to view this content.
Results
This protocol performs immunofluorescence of LC3 in pancreatic cell lines to determine the autophagy levels in different conditions. The outcome of this experiment was the obtention of cellular images from the red and blue channels, corresponding to LC3 and DAPI. The LC3 images indicate the cellular distribution of this protein, whereas the DAPI shows the nuclear localization. Figure 2A shows a representative image of the immunofluorescence of LC3 and its merge with DAPI staining in PANC-1 c...
Access restricted. Please log in or start a trial to view this content.
Discussion
The method described in this protocol allows for visualizing the endogenous LC3 distribution in the cell and quantifying the autophagic levels under different conditions. Another similar method used to analyze the LC3 distribution and determine autophagy activation involves fluorescence-labeled LC3 transfection (such as RFP-LC3)19. RFP-LC3 transfection has the advantages of not needing fixation (which allows for applying this method in live cell imaging20), being cheaper, a...
Access restricted. Please log in or start a trial to view this content.
Disclosures
No conflicts of interest were declared.
Acknowledgements
This work was supported by grants from the University of Buenos Aires (UBACyT 2018-2020 20020170100082BA), the National Council for Scientific Research and Technology (CONICET) (PIP 2021-2023 GI− 11220200101549CO; and PUE 22920170100033) and the National Agency for Scientific and Technological Promotion (PICT 2019-01664).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 10x Phosphate-Buffered Saline (PBS) | Corning | 46-013-CM | |
| 12 mm round coverslips | HDA | CBR_OBJ_6467 | |
| 24 Well- Cell Culture Plate | Sorfa | 220300 | |
| Absolute ethanol | Biopack | 2207.10.00 | |
| Alexa Fluor 594 Donkey anti-rabbit IgG (H+L) | Invitrogen | R37119 | |
| Confocal Laser Scanning Microscope | Zeiss | LSM 800 | |
| Dexamethasone | Sigma Aldrich | D4902 | |
| DMEN | Sartorius | 01-052-1A | |
| Fetal Bovine Serum | NATOCOR | Lintc-634 | |
| Gemcitabina | Eli Lilly | VL7502 | |
| LC3B (D11) XP Rabbit mAb | Cell Signaling Technology | 3868S | |
| Methanol | Anedra | 6197 | |
| Parafilm "M" (Laboratory Sealing Film) | Bemis/Curwood | PM-996 | |
| Pen-Strep Solution | Sartorius | 03-031-1B | |
| PP242 | Santa Cruz Biotechnology | SC-301606 | |
| Trypsin EDTA | Gibco | 11570626 |
References
- Grasso, D., Renna, F. J., Vaccaro, M. I. Initial steps in mammalian autophagosome biogenesis. Frontiers in Cell and Developmental Biology. 6, 146(2018).
- Galluzzi, L., et al. Molecular definitions of autophagy and related processes. EMBO Journal. 36 (13), 1811-1836 (2017).
- Kitada, M., Koya, D. Autophagy in metabolic disease and ageing. Nature Reviews Endocrinology. 17 (11), 647-661 (2021).
- Ktistakis, N. T., Tooze, S. A. Digesting the expanding mechanisms of autophagy. Trends in Cell Biology. 26 (8), 624-635 (2016).
- Tanida, I., Ueno, T., Kominami, E. LC3 and autophagy. Methods in Molecular Biology. 445, 77-88 (2008).
- Kabeya, Y., et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO Journal. 19 (21), 5720-5728 (2000).
- Mizushima, N., Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy. 3 (6), 542-545 (2007).
- Vanasco, V., et al. Mitochondrial dynamics and VMP1-related selective mitophagy in experimental acute pancreatitis. Frontiers in Cell and Developmental Biology. 9, 640094(2021).
- Williams, J. A. Regulatory mechanisms in pancreas and salivary acini. Annual Review of Physiology. 46, 361-375 (1984).
- Mizrahi, J. D., Surana, R., Valle, J. W., Shroff, R. T. Pancreatic cancer. Lancet. 395 (10242), 2008-2020 (2020).
- Li, J., et al. Regulation and function of autophagy in pancreatic cancer. Autophagy. 17 (11), 3275-3296 (2021).
- Ropolo, A., et al. A novel E2F1-EP300-VMP1 pathway mediates gemcitabine-induced autophagy in pancreatic cancer cells carrying oncogenic KRAS. Frontiers in Endocrinology. 11, 411(2020).
- Pardo, R., et al. Gemcitabine induces the VMP1-mediated autophagy pathway to promote apoptotic death in human pancreatic cancer cells. Pancreatology. 10 (1), 19-26 (2010).
- Logsdon, C. D., Moessner, J., Williams, J. A., Goldfine, I. D. Glucocorticoids increase amylase mRNA levels, secretory organelles, and secretion in pancreatic acinar AR42J cells. Journal of Cell Biology. 100 (4), 1200-1208 (1985).
- Klionsky, D. J., et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy. 17 (1), 1(2021).
- Segeritz, C. -P., Vallier, L. Chapter 9 - Cell culture: Growing cells as model systems in vitro. Basic Science Methods for Clinical Researchers. Jalali, M., Saldanha, F. Y. L., Jalali, M. , Academic Press. Cambridge, MA. 151-172 (2017).
- Elliott, A. D. Confocal microscopy: Principles and modern practices. Current Protocols in Cytometry. 92 (1), 68(2020).
- Grasso, D., et al. a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. Journal of Biological Chemistry. 286 (10), 8308-8324 (2011).
- Ropolo, A., et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. Journal of Biological Chemistry. 282 (51), 37124-37133 (2007).
- Karanasios, E., Stapleton, E., Walker, S. A., Manifava, M., Ktistakis, N. T. Live cell imaging of early autophagy events: Omegasomes and beyond. Journal of Visualized Experiments. (77), e50484(2013).
- Kuma, A., Matsui, M., Mizushima, N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 3 (4), 323-328 (2007).
- Zhang, Z., Singh, R., Aschner, M. Methods for the detection of autophagy in mammalian cells. Current Protocols in Toxicology. 69, 1-26 (2016).
- Yoshii, S. R., Mizushima, N. Monitoring and measuring autophagy. International Journal of Molecular Sciences. 18 (9), 1865(2017).
- Betriu, N., Andreeva, A., Semino, C. E. Erlotinib promotes ligand-induced EGFR degradation in 3D but not 2D cultures of pancreatic ductal adenocarcinoma cells. Cancers. 13 (18), 4504(2021).
- Wang, W., Dong, L., Zhao, B., Lu, J., Zhao, Y. E-cadherin is downregulated by microenvironmental changes in pancreatic cancer and induces EMT. Oncology Reports. 40 (3), 1641-1649 (2018).
- Kim, S. K., et al. Phenotypic heterogeneity and plasticity of cancer cell migration in a pancreatic tumor three-dimensional culture model. Cancers. 12 (5), 1305(2020).
- Meng, Y., et al. Cytoplasmic EpCAM over-expression is associated with favorable clinical outcomes in pancreatic cancer patients with Hepatitis B virus negative infection. International Journal of Clinical and Experimental Medicine. 8 (12), 22204-22216 (2015).
- Brock, R., Hamelers, I. H., Jovin, T. M. Comparison of fixation protocols for adherent cultured cells applied to a GFP fusion protein of the epidermal growth factor receptor. Cytometry. 35 (4), 353-362 (1999).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved