A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
An Antimicrobial Fabric Using Nano-Herbal Encapsulation of Essential Oils
In This Article
Summary
Antimicrobial lab coats prevent the cross-contamination of pathogen accumulation and accidental bio-spills. Here, we describe the protocol for developing a skin-friendly antimicrobial fabric using nano-herbal encapsulation and modified standard tests to precisely evaluate the efficacy and suitability for typical usage of the lab coat.
Abstract
Lab coats are widely used in biohazard laboratories and healthcare facilities as protective garments to prevent direct exposure to pathogens, spills, and burns. These cotton-based protective coats provide ideal conditions for microbial growth and attachment sites due to their porous nature, moisture-holding capacity, and retention of warmth from the user's body. Several studies have demonstrated the survival of pathogenic bacteria on hospital garments and lab coats, acting as vectors of microbial transmission.
A common approach to fix these problems is the application of antimicrobial agents in textile finishing, but concerns have been raised due to the toxicity and environmental effects of many synthetic chemicals. The ongoing pandemic has also opened a window for the investigation of effective antimicrobials and eco-friendly and toxic-free formulations. This study uses two natural bioactive compounds, carvacrol and thymol, encapsulated in chitosan nanoparticles, which guarantee effective protection against four human pathogens with up to a 4-log reduction (99.99%). These pathogens are frequently detected in lab coats used in biohazard laboratories.
The treated fabrics also resisted up to 10 wash cycles with 90% microbial reduction, which is sufficient for the intended use. We made modifications to the existing standard fabric tests to better represent the typical scenarios of lab coat usage. These refinements allow for a more accurate evaluation of the effectiveness of antimicrobial lab coats and for the simulation of the fate of any accidental microbial spills that must be neutralized within a short time. Further studies are recommended to investigate the accumulation of pathogens over time on antimicrobial lab coats compared to regular protective coats.
Introduction
The protective white coat is a mandatory personal protective equipment (PPE) item in microbiology laboratories and healthcare facilities, and it protects from direct exposure to pathogens, spills, and burns. These cotton coats promote microbial growth due to many factors-the woven fabric provides attachment sites and aeration, cotton and starch used in the manufacturing process along with exfoliated epithelial cells from the user supply nutrients, and the proximity to the user gives warmth and moisture. The accumulation of microbes on textiles can also cause health problems such as allergies and nosocomial infection, unpleasant odors, and fabric deterioration1.
Unlike regular clothing, protective coats are infrequently washed or disinfected, as found in many surveys2,3. Many studies show evidence of lab coats acting as a vector of microbial transmission and the risk of nosocomial infections in the healthcare setting2,4, particularly resistant strains3 such as methicillin-resistant Staphylococcus aureus (MRSA); thus, they raise health concerns of PPE, which is meant to protect from microbial contamination. There are not enough cross-sectional studies on lab coat-associated infections in the context of Biosafety Level 2 (BSL-2) facilities or microbiology teaching labs, but many regulatory authorities restrict the use of lab coats within the containment level. However, many academic institutions in North America struggle to meet the requirements due to practical constraints, such as laundering and storing inside the facility, the incidents of wearing lab coats in public areas such as cafeterias and libraries are common. One practical solution to these issues is the application of antimicrobial agents in textile finishing.
Antimicrobial fabrics are gaining increasing popularity in sportswear, activewear, and socks, mainly intended to reduce body odor. However, the use of these fabrics is not common in PPE development, except for some silver-coated cotton masks and healthcare garments5. We report the development of an antimicrobial fabric for lab coats, which inhibits common pathogens found in BSL-2 labs and renders effective protection from the cross-contamination of common pathogens.
Currently, a variety of antimicrobial fabrics and finishings are available in the market, but most of these use heavy metal colloidal particles (e.g., silver, copper, zinc), organometallics, or synthetic chemicals such as triclosan and quaternary ammonium compounds, which are not environmentally friendly1 and may lead to health issues such as skin irritation and allergies6. Some synthetic formulations pose concerns due to non-target microbes, such as normal flora or inducing antimicrobial resistance (AMR). The US Food and Drug Administration (FDA) regulates commercial antimicrobial fabrics, which must be non-toxic to the user and free from eco-toxicity. Therefore, antimicrobial fabrics based on natural biocides that inhibit a broad spectrum of microbes are preferable. Essential oils (EOs) are used widely as antimicrobial and therapeutic agents, but their use in antimicrobial finishing is limited due to their durability6,7,8. Based on our knowledge and market research on nano-herbal finishing8, no herbal-based antimicrobial fabric is commercially available. This is because synthetic coatings are easy to manufacture and have long durability. A few nano-herbal-coated textiles reported only for research purposes include neem7, moringa9, and curry leaves9.
The present study uses two bioactive components extracted from oregano EOs, carvacrol and thymol, which are effective against a wide range of bacterial pathogens and viruses but are generally recognized as safe for humans10. However, these bioactive components are volatile, and therefore their antimicrobial potential is short-lived if applied directly to the fabric. Nano-herbal encapsulation is a process in which bioactive components or drugs are loaded inside a polymeric shell that protects the core from environmental degradation, and thus enhances the shelf life. In addition, the small size of the polymeric particles, which generally range from 10 nm to 100 nm, enhances the efficacy of the application and slows the release of the bioactive compounds onto the fabric. These bioactive compounds are used for various purposes, such as food preservation10, but not for textile coating.
Among many polymeric encapsulants, chitosan is an attractive candidate due to many of its attributes, such as nontoxicity, biodegradability, mucoadhesivity, and biocompatibility11. It is a natural polysaccharide, obtained by the deacetylation process from chitin, which is found in seashells and fungal cell walls. It is used in biochemical and food preservation applications such as drug or protein delivery11,12,13, controlled release14, and antimicrobial films10. Chitosan is not readily soluble in water but forms a colloidal suspension in acidic media. Bioactive molecules are loaded into chitosan nanoparticles (NPs) by a simple two-step ionic gelation method14,15,16. In this process, hydrophobic bioactive compounds such as carvacrol and thymol form an oil-in-water emulsion, which is aided by a surfactant, Tween 80. Subsequently, a polyanionic compound, pentasodium tripolyphosphate (TPP), is used to form the cross-linkages between the amino groups along the polycationic polymer molecules and phosphate groups of TPP molecules to stabilize the complex. This complexation process solidifies the bioactive compounds within the matrix of chitosan, which is subsequently purified and coated onto cotton swatches to produce antimicrobial fabric.
The nano-formulations must be tested first for antimicrobial effectiveness in emulsion form before being applied to the fabric. This can be conveniently evaluated by a qualitative method, such as Kirby-Bauer disk diffusion, well diffusion, and the cylinder plate assay. However, the cylinder plate assay17 provides the flexibility to load varying volumes of the formulation and compare the zone of clearance. In this method, the antimicrobial formulations are loaded in stainless steel cylinders and placed on a soft agar layer, which is inoculated with the test microorganism or pathogen. The diameter of the zone of clearance produced against the test organism is proportional to the inhibitory potential of the antimicrobial formulation, and therefore can be used as an alternative to broth dilution methods. However, the size of the clear zones is only a comparative or qualitative measure within a specific plate unless specific standards are maintained. Antimicrobial agents act against the pathogens either by inhibiting their growth (biostatic) or killing the cells (biocidal), which can be quantified by minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC), respectively. However, the efficacy and behavior of the bioactive chemicals are different in their formulations (liquid state) and when coated on a substrate such as a fabric18. This is because multiple factors play a role in the efficacy, such as the stability of the adherence of the antimicrobial agents to the fabric, moisture content, substrate type, and adherence of the microbes. If the intended purpose is only bacteriostatic activity, a qualitative assay such as the "Parallel Streak Method"19 can provide a relatively quick and easy evaluation of diffusible antimicrobial formulation. However, if the bactericidal effects are to be determined, "Assessment of Antibacterial Finishes on Textile Materials"20 can be employed, which provides the log reduction of the spiked pathogen.
Protocol
1. Preparation of nanoparticles
- Nano-herbal encapsulation
- Prepare 50 mL of 1% (v/v) acetic acid.
CAUTION: Glacial acetic acid is an irritant, which can cause severe skin burns and eye damage. Wear a full-length lab coat, nitrile gloves, and goggles, and work under a fume hood. - Prepare chitosan solution (1.2% w/v) by dissolving 0.6 g of chitosan flakes (medium molecular weight) in 50 mL of 1% acetic acid (prepared above). Agitate overnight (O/N) at room temperature (R/T) to get a homogeneous emulsion.
- Add 0.5 g of Tween 80 and stir (1,000 rpm) at 60 °C for 2 h to get a homogeneous solution. Bring the solution to R/T before adding bioactive compounds (carvacrol or thymol).
- Add 0.75 g of carvacrol (or thymol) dropwise or gradually while stirring (1,000 rpm) the mixture for 20 min at R/T. The weight ratio of chitosan to bioactive compound is 1:1.25.
- Add 50 mL (0.5% w/v) of TPP dropwise to the mixture while stirring at R/T. Continue stirring for 30 min to get a homogeneous emulsion.
- Prepare a negative control by following the same procedure but without the addition of bioactive compounds.
- Prepare 50 mL of 1% (v/v) acetic acid.
- Purification
- Centrifuge the emulsion at 10,000 × g for 30 min at 4 °C and collect the formed NPs (pellet) after decanting the supernatant. Reserve the supernatant to test the encapsulation efficacy.
- Wash the particles (from step 1.2.1) using aqueous Tween 80 (1% v/v) with double the volume of pellets formed to remove the unbound or free bioactive compounds. Make sure to disturb the pellet by vortexing until a homogeneous solution is formed in each wash step.
- Wash the particles (from step 1.2.2) with deionized water twice to get rid of impurities.
- Reconstitute the NPs by resuspending the pellet (from step 1.2.3) in 30 mL of deionized water. Store the NPs at 4 °C for up to 6 months.
NOTE: The experiment can be paused at this stage.
- Characterization
- Dilute the NPs in deionized water until the ultraviolet-visible (UV-Vis) readings between 250 to 400 nm fall within the range (absorbance >1).
- Record the UV-Vis absorption spectra of the NPs over the wavelengths ranging from 250 to 400 nm using a UV-Vis spectrophotometer.
- Store an aliquot (1 mL) of NPs at R/T for a range of durations (e.g., 1 to 6 months) and test the antimicrobial effect by the cylinder plate method along with a fresh sample.
- Coating on fabric
- Apply NPs on a woven cotton fabric using a pad-dry-cure method7,21.
- Immerse the fabric swatches in the solution containing NPs mixed with a fabric binder for 3 min until soaked. Then, pass the swatches through a two-roller laboratory padder to remove excess liquid.
- Oven-dry the swatches at 100 °C for 30 min and cure at 130 °C for 10 min.
- Finally, wash the swatches in an ultrasound bath for 15 min to remove unbound NPs.
NOTE: Alternatively, follow the simplified method described below.
- Cut the cotton fabric (or lab coat swatches) into 10 cm x 10 cm squares.
- Immerse the cut pieces in 2 mL of the synthesized NP (10%) in a plastic container (12 cm x 12 cm) for 2 min at R/T. Remove excess liquid by air drying.
- Heat press at 100 °C for 3 min to bind the NPs.
- Remove unbound NPs by rinsing in aqueous Tween 80 (2% w/v) and dry in the oven at 100 °C for 30 min.
NOTE: This antimicrobial fabric can be stored for 2 years at R/T.
- Apply NPs on a woven cotton fabric using a pad-dry-cure method7,21.
- Wash durability
- Cut the fabric into 50 mm x 50 mm square swatches.
- Place the swatches in 50 mL of warm tap water (40 °C) in a glass/plastic container and add two drops of regular detergent (non-antimicrobial).
- Rinse on a rocking platform shaker or magnetic stirrer for 30 min.
- Air dry or incubate at 60 °C for 2 h and test for antimicrobial efficacy.
2. Cylinder plate assay for screening of nanoparticles
- Prepare trypticase soy agar (TSA), trypticase soy broth (TSB), antibiotic base agar (ABA), and antibiotic seed agar (ASA), according to manufacturers' instructions, and sterilize the media by autoclaving at 121 °C at 15 psi for 15 min.
- Subculture the microbes (from stock) of interest, against which the efficacy tests are performed, on freshly prepared TSA plates (or any appropriate media) by the three-way streak plate method and incubate to produce purity plates. (Recommended species: S. aureus, E. coli, P. aeruginosa, and C. albicans.)
- Inoculate the cultures from fresh purity plates in TSB (10 mL tubes) at 35 °C O/N with moderate shaking (100-200 rpm).
- Aliquot 5.0 mL of the molten ASA in each of the test tubes. The number of tubes corresponds to the number of microorganisms tested.
- Pour 20 mL of presterilized molten ABA into each Petri plate (100 mm x 20 mm) aseptically to form the base layer. The number of agar plates corresponds to the number of microorganisms to be tested. Wait until the media get solidified.
- After solidification of the base layer, inoculate each molten ASA with an overnight culture from step 2.3 (1.0 mL, prewarmed to 35 °C) and immediately transfer the contents (mixture of ASA and culture) to the surface of an ABA plate. Swirl the plate to distribute the molten agar layer evenly. Ensure that the seed layer appears smooth and free from bumps or bubbles.
- Place up to six stainless steel cylinders (6 mm x 6 mm x 10 mm; preautoclaved), evenly spaced on a hexagonal pattern, per agar plate using a pair of sterile tweezers (Figure 1).
- Load the cylinders with the synthesized NPs of different volumes (30 µL, 50 µL, 75 µL, and 100 µL) to be screened for antimicrobial effects. The negative control is the one without any bioactive compounds.
- Incubate common bacterial pathogens (e.g., E. coli, S. aureus, P. aeruginosa) at 35 °C for 24 h and fungal species (e.g., C. albicans) at R/T for 3 days.
- Measure the diameter of the clear zone and compare the effectiveness of the synthesized NPs.
NOTE: This test can be used to prescreen the best NPs to be coated on the fabric.
3. Parallel streak method (modified from AATCC 147)
- Material preparation
- Cut the NP-treated fabric into 50 mm x 25 mm swatches.
- Prepare Mueller-Hinton agar (MHA), TSA, and TSB, according to manufacturers' instructions, and sterilize the media by autoclaving at 121 °C at 15 psi for 15 min.
- Subculture the microbes (from stock) of interest, against which the efficacy tests are performed, on freshly prepared TSA plates (or any appropriate media) by the three-way streak plate method and incubate at 35 °C for 2 days for bacterial stains and at R/T for 5 days for fungal strains to produce purity plates. (Recommended species: S. aureus, E. coli, P. aeruginosa, and C. albicans.)
- Spiking of microbial cultures
- Inoculate the cultures from fresh purity plates in TSB (10 mL tubes) at 35 °C O/N with moderate shaking (100-200 rpm).
- Dilute the O/N cultures to 1.5 × 108 colony forming units (CFU)/mL or corresponding to the turbidity of 0.5 McFarland standard (usually a 1/10 to 1/20 dilution for most healthy cultures).
- Inoculate the diluted culture above using a 4 mm sterile loop as follows. Load a loopful of broth culture and transfer it to the surface of the MHA agar plate by making five parallel streaks, each 6 cm in length and 1 cm apart, according to Figure 2. Do not refill the loop.
- Gently press the fabric swatch using a sterile spatula across the five streaks so that the fabric is in the center and touches all five streak lines. Incubate at 35 °C for 24 h.
- Qualitative evaluation of antimicrobial efficacy
- Examine the incubated plate for the interruption of growth along the streaks beyond the edges of the fabric (clear zone indicates inhibition of growth).
- Calculate the average width of a zone of inhibition along the streak line (W) on either side of the fabric swatch using equation (1).
W = (T - D)/2 (1)
W = width of clear zone; T = total length of the clear zone, including the swatch width; D = width of the fabric swatch (25 mm).
4. Quantitative log reduction method (modified from AATCC 100)
- Experimental preparation
- Cut the fabric into 50 mm x 50 mm square swatches.
- Prepare TSA, TSB, Letheen broth, and phosphate-buffered saline (PBS), according to manufacturers' instructions, and sterilize the media by autoclaving.
- Prepare O/N cultures of microbes of interest, against which the efficacy tests are performed, by inoculating isolated colonies from purity plates in sterile TSB and incubating at 35 °C for 18-24 h. (Recommended species: S. aureus, E. coli, P. aeruginosa, and C. albicans.)
- Spiking of microbial cultures
- Dilute the O/N cultures to 1.5 × 108 CFU/mL or corresponding to the turbidity of 0.5 McFarland standard (usually a 1/10 to 1/20 dilution for most healthy cultures).
- Determine the appropriate volume of cultures for spiking by measuring the liquid holding capacity of the fabric as follows.
- Add a series of volumes of the diluted broth culture (e.g., 100-500 µL) onto fabric swatches (in separate Petri plates) and choose the volume such that the fabric swatch absorbs the water fully and leaves no residual/free liquid. The liquid holding capacity differs from the type and thickness of the fabric. For regular cotton lab coats, it is roughly 200 µL.
- Spike the volume (liquid-holding capacity determined above [e.g., 200 µL]) of each culture onto the swatches placed in sterile Petri plates. The number of swatches corresponds to the number of tests (e.g., fabric tested immediately and after washing [and the laundering cycles tested]), and the antimicrobial effects tested over the contact time (after a set time/day [e.g., 30 min, 2 h, Day 1-3]).
NOTE: Use a micropipette with aerosol filter tips (to prevent contamination of the pipette in subsequent uses) to inoculate the cultures on the swatches with even distribution. - For the negative control, use the same type of untreated fabric swatches. Perform the negative control for each corresponding test-microbial species, contact period, different fabrics, and wash cycles.
- Recovery of microbes by viable plate counts
- Allow the inoculated swatches (both treated and untreated) to air-dry inside the Petri plates (lids ajar) at R/T for the required contact period/s to be tested (e.g., 0 min, 30 min, 60 min, etc., or even days for long-term effects). Always include a "0 min" to represent an immediate effect and neutralizing efficacy.
- Transfer the swatches aseptically to separate sterile centrifuge tubes (50 mL) and screw the caps tightly.
- Add Letheen broth (any relevant neutralizing buffers) to make a 1/100 dilution (e.g., 19.8 mL for an inoculum of 200 µL).
- Close the centrifuge tubes with screw caps tightly and vortex for 1 min at medium speed.
- Serially dilute the suspension with the sterile PBS in subsequent 1/10 dilutions, such that the colony counts of the "untreated" group becomes too low to count (TLTC).
- Plate the dilutions (0.1 mL) on appropriate media plates, which support the growth of the microorganism (e.g., TSA for bacteria or Sabouraud dextrose agar [SDA] for fungi) or any media that optimize the growth and provide good contrast to make the colony counting accurate.
- Incubate the bacterial plates at 35 °C for 2 days and the fungal plates at R/T for 5 days.
- Count the viable CFUs directly using a colony counter or using imaging software (e.g., CFU AI).
- Calculate the microbial log reduction (R) due to antimicrobial fabric using equation (2):
R =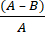 × 100 (2)
× 100 (2)
A = log value of the number of CFUs recovered from untreated fabric; B = log value of the number of CFUs recovered from treated fabric.
Results
Initial screening of the synthesized NPs
Following the two-step oil-in-water emulsion technique16, the bioactive compounds (carvacrol and thymol) were successfully encapsulated in chitosan. This was confirmed by UV-Vis spectrophotometry for the peak absorption of the respective bioactive compounds compared to controls, which were the chitosan NPs without any bioactive compounds. The constituted NPs were homogeneous and stable over 12 months at 4 °C. The initial screenin...
Discussion
The antimicrobial efficacy of biocides is conventionally tested by quantitative assays, such as minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC), in which the bacteria are immersed in an antimicrobial liquid for 24 h. However, these assays are not suitable for coated fabrics, where the liquid interface is lacking and the biocides are diffused slowly along the fabric fibers. Therefore, many standard fabric tests have been established, such as AATCC 147, ISO 20645, AATCC 100, and JIS L 19...
Disclosures
The authors have no conflicts of interests.
Acknowledgements
This study was funded by "Applied Research, Innovation and Entrepreneurship Services" (ARIES), Centennial College, Canada.
Materials
| Name | Company | Catalog Number | Comments |
| Acetic acid | Millipore Sigma | 64-19-7 | |
| Antibiotic base agar | BD Difco | DF0270-17-4 | Also known as Antibiotic Medium 2 |
| Antibiotic seed agar | BD Difco | DF0263-17-3 | Also known as Antibiotic Medium 1 |
| Blood Agar (Nutrient Agar with 5% Sheep Blood) | Donated by CFIA | ||
| Bromcresol Purple Lactose Agar | Donated by CFIA | ||
| Candida albicans | ATCC The Global Bioresource Center | ATTC 10231 | |
| Carvacrol | Millipore Sigma | 282197 (CAS# 499-75-2) | |
| Centrifuge Allergra X-22R Centrifuge | Beckman Coulter | Model # X-22R | Refrigerated. Wait at least 20 min or until the temperature reach the set low value (e.g., 4 °C) as the refrigeration takes time. |
| Chitosan Medium Molecular Weight (CS) | Millipore Sigma | 448877 (CAS # 9012-76-4) | |
| Clamshell Heat Press | Intiva | IM1200 | |
| Escherichia coli (E. coli) | ATCC The Global Bioresource Center | ATTC 23725 | |
| Incubator | Thermo Scientific | 1205M34 | |
| Letheen Broth | BD Difco | DF0681-17-7 | Used to neutralize antimicrobial effects. Product from different manufacturers may require to add Polysorbate 80, which is already added in Difco product. |
| Milli Q water | Millipore Sigma | ZR0Q16WW | Deionized water |
| Mueller-Hinton Agar | BD Difco | DF0252-17-6 | |
| Pentasodium tripolyphosphate (TPP) | Millipore Sigma | 238503 (CAS# 7758-29-4) | |
| Phospahte Buffered Saline (PBS) | Thermo Scientific | AM9624 | |
| Pseudomonas aeruginosa | ATCC The Global Bioresource Center | ATTC 9027 | |
| Sabouraud Dextrose Agar | BD Difco | DF0109-17-1 | |
| Shaking incubator/ Thermo shaker | VWR | Model# SHKA2000 | |
| Staphylococcus aureus | ATCC The Global Bioresource Center | ATTC 6538 | |
| Thymol | Millipore Sigma | T0501 (CAS# 89-83-8) | |
| Trypticase Soy Agar | BD Difco | 236950 | |
| Trypticase Soy Broth | BD Difco | 215235 | |
| Tween 80 | Millipore Sigma | STS0204 (CAS # 9005-65-6) | |
| UV-Vis Spectrophometer | Thermo Scientific | GENESYS 30 (840-277000) |
References
- Schmidt-Emrich, S., et al. Rapid assay to assess bacterial adhesion on textiles. Materials. 9 (4), 249 (2016).
- Qaday, J., et al. Bacterial contamination of medical doctors and students white coats at Kilimanjaro Christian Medical Centre, Moshi, Tanzania. International Journal of Bacteriology. 2015, 507890 (2015).
- Treakle, A. M., et al. Bacterial contamination of health care workers' white coats. American Journal of Infection Control. 37 (2), 101-105 (2009).
- Wong, D., Nye, K., Hollis, P. Microbial flora on doctors' white coats. BMJ. 303 (6817), 1602-1604 (1991).
- Gouveia, I. C. Nanobiotechnology: A new strategy to develop non-toxic antimicrobial textiles for healthcare applications. Journal of Biotechnology. (150), 349 (2010).
- Joshi, M., Ali, S. W., Purwar, R., Rajendran, S. Ecofriendly antimicrobial finishing of textiles using bioactive agents based on natural products. Indian Journal of Fibre and Textile Research. 34, 295-304 (2009).
- Ahmed, H. A., Rajendran, R., Balakumar, C. Nanoherbal coating of cotton fabric to enhance antimicrobial durability. Elixir Applied Chemistry. 45, 7840-7843 (2012).
- Morais, D. S., Guedes, R. M., Lopes, M. A. Antimicrobial approaches for textiles: From research to market. Materials. 9 (6), 498 (2016).
- Venkatraman, P. D., Sayed, U., Parte, S., Korgaonkar, S. Development of advanced textile finishes using nano-emulsions from herbal extracts for organic cotton fabrics. Coatings. 11 (8), 939 (2021).
- Martínez-Hernández, G. B., Amodio, M. L., Colelli, G. Carvacrol-loaded chitosan nanoparticles maintain quality of fresh-cut carrots. Innovative Food Science & Emerging Technologies. 41, 56-63 (2017).
- Zhang, H. L., Wu, S. H., Tao, Y., Zang, L. Q., Su, Z. Q. Preparation and characterization of water-soluble chitosan nanoparticles as protein delivery system. Journal of Nanomaterials. 2010, 1-5 (2010).
- Patel, R., Gajra, B., Parikh, R. H., Patel, G. Ganciclovir loaded chitosan nanoparticles: preparation and characterization. Journal of Nanomedicine & Nanotechnology. 7 (6), 1-8 (2016).
- Merodio, M., Arnedo, A., Renedo, M. J., Irache, J. M. Ganciclovir-loaded albumin nanoparticles: characterization and in vitro release properties. European Journal of Pharmaceutical Sciences. 12 (3), 251-259 (2001).
- Hsieh, W. C., Chang, C. P., Gao, Y. L. Controlled release properties of Chitosan encapsulated volatile Citronella Oil microcapsules by thermal treatments. Colloids and Surfaces B: Biointerfaces. 53 (2), 209-214 (2006).
- Yoksan, R., Jirawutthiwongchai, J., Arpo, K. Encapsulation of ascorbyl palmitate in chitosan nanoparticles by oil-in-water emulsion and ionic gelation processes. Colloids and Surfaces B: Biointerfaces. 76 (1), 292-297 (2010).
- Keawchaoon, L., Yoksan, R. Preparation, characterization and in vitro release study of carvacrol-loaded chitosan nanoparticles. Colloids and Surfaces B: Biointerfaces. 84 (1), 163-171 (2011).
- Cazedey, E. C. L., Salgado, H. R. N. Development and validation of a microbiological agar assay for determination of orbifloxacin in pharmaceutical preparations. Pharmaceutics. 3 (3), 572-581 (2011).
- Jayapriya, S., Bagyalakshmi, G. Textile antimicrobial testing and standards. International Journal of Textile and Fashion Technology. 4 (1), 2250-2378 (2013).
- AATCC 100. Antibacterial Finishes on Textile Materials: Assessment of Developed from American Association of Textile Chemists and Colorists. AATCC 100. , (2004).
- AATCC 147. Antimicrobial Activity Assessment of Textile Materials: Parallel Streak Method from American Association of Textile Chemists and Colorists. AATCC 147. , (2004).
- Ortelli, S., Costa, A. L., Dondi, M. TiO2 nanosols applied directly on textiles using different purification treatments. Materials. 8 (11), 7988-7996 (2015).
- Poole, K. Pseudomonas aeruginosa: resistance to the max. Frontiers in Microbiology. 2, 65 (2011).
- Pinho, E., Magalhães, L., Henriques, M., Oliveira, R. Antimicrobial activity assessment of textiles: standard methods comparison. Annals of Microbiology. 61 (3), 493-498 (2010).
- Venkatraman, P. D., Sayed, U., Parte, S., Korgaonkar, S. Novel antimicrobial finishing of organic cotton fabrics using nano-emulsions derived from Karanja and Gokhru plants. Textile Research Journal. 92 (23-24), 5015-5032 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved