A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Detecting SARS-CoV-2 Virus by Reverse Transcription-Loop-Mediated Isothermal Amplification
In This Article
Summary
Here we provide a complete protocol to standardize and implement the method for detecting the SARS-CoV-2 virus in human samples by reverse transcription loop-mediated isothermal amplification (RT-LAMP). This method, done within 60 min, could be adapted to any laboratory or point-of-care at a low cost and using inexpensive equipment.
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has dramatically impacted human health. It continues to be a threat to modern society because many people die as a result of infection. The disease is diagnosed using serologic and molecular tests, such as the gold standard real-time polymerase chain reaction (RT-PCR). The last has several disadvantages because it requires specialized infrastructure, costly equipment, and trained personnel. Here, we present a protocol outlining the steps required to detect the SARS-CoV-2 virus using reverse transcription-loop-mediated isothermal amplification (RT-LAMP) in human samples. The protocol includes instructions for designing primers in silico, preparing reagents, amplification, and visualization. Once standardized, this method can be easily implemented and adapted to any laboratory or point-of-care within 60 min at a low cost and using inexpensive equipment. It is adaptable to detecting different pathogens. Thus, it can potentially be used in the field and in health centers to carry out timely epidemiological surveillance.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19). The World Health Organization declared a public health emergency of international concern on 30 January 2020 and a pandemic on 11 March 2020. The pandemic resulted in over 760 million cases and 6.87 million deaths as of the date this article was written1.
The impact of this virus has highlighted the need for better, more accurate, faster, and more widely available surveillance tools to improve infectious disease detection and control2,3. During the pandemic, SARS-CoV-2 diagnostic tests were based on detecting nucleic acid, antibodies, and proteins, but RT-PCR detection of nucleic acid is the gold standard4. However, RT-PCR has some limitations; it requires specialized equipment, infrastructure, and personnel trained in molecular biology, limiting its application to specialized laboratories. Further, it is time-consuming (4-6 h), not including the time to transport the specimens to the laboratory, which can take days5. These constraints prevent efficient sample processing and obtaining the information required for contingency planning and epidemiological management.
Reverse transcription-loop-mediated isothermal amplification (RT-LAMP) has several advantages over RT-PCR, making it an appealing strategy for designing future point-of-care diagnostic tests (POCT), particularly in resource-constrained settings6. First, it is greatly specific because it uses between four and six primers that recognize six to eight areas in the target sequence, be it DNA or RNA7,8. Second, because it operates at a constant temperature, it does not require sophisticated equipment such as real-time thermal cyclers to generate the amplification, nor does it necessitate highly trained personnel to operate it. Third, the reaction time is very short (~60 min), and reagents that are not very specialized are employed, which makes it a cost-effective tool6. Given the foregoing and the health emergency caused by the COVID-19 pandemic, this technique can be viewed as an alternative diagnostic method that is quick, inexpensive, and simple to implement in any research laboratory9.
The protocol for standardizing and implementing an RT-LAMP to detect SARS-CoV-2 by colorimetric methods using a thermocycler and a water bath is described in this article (Figure 1). Critical points, their limitations, and alternatives to advance them are discussed.
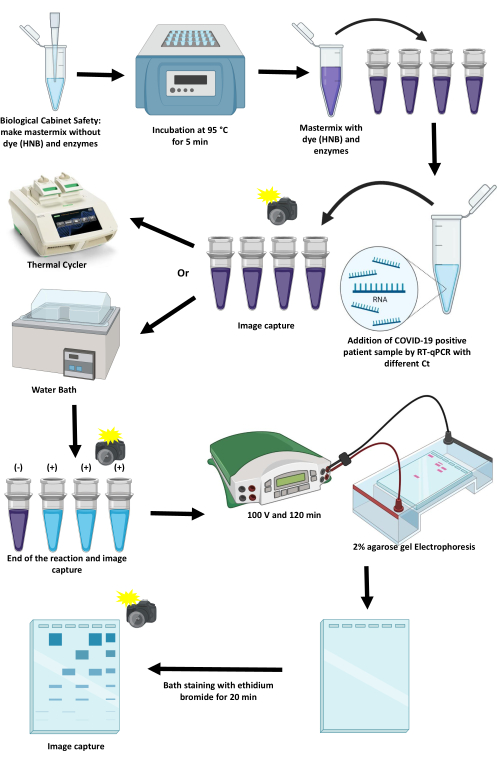
Figure 1: Scheme of the protocol for amplifying SARS-CoV-2 using the RT-LAMP technique. Please click here to view a larger version of this figure.
Protocol
The samples used were provided by the clinical laboratory of Fundación Valle del Lili University hospital and corresponded to the purified RNA from patients who tested positive for COVID-19 using the RT-qPCR technique. All patients provided informed consent for research, and this study was approved by the bioethical committee for human studies from Fundación Valle del Lili University hospital.
1. RT-LAMP primer design and preparation
NOTE: LAMP primers can be used with a variety of platforms, including New England BioLabs (NEB) LAMP, Primer Explorer, and LAMP assay versatile analysis (LAVA). However, for this protocol, the NEB LAMP tool was used. Primer design can be done using SARS-CoV-2 genomes obtained from NextStrain database10. Table 1 shows the primer set used in this protocol.
- Primer design for LAMP
- Obtain viral genome sequences.
- Perform sequence alignments to obtain the consensus sequence.
- Navigate to the NEB LAMP primer design tool platform11 and follow the instructions in the quick guide. This tool produces the same results as primer explorer V5, but it is much more user-friendly in its output. Use primer explorer user manuals as a guide for primer design.
- Thermodynamic evaluation of the set of primers
- Use the tool Primer-Dimer12 to perform a thermodynamic analysis on the primers obtained.
- Put the primer sequences in the tool. Then, select the option Multiplex Analysis and Dimer Structure Report.
- Select the primer sets that have ΔG not less than -5.
- Specificity evaluation of the designed primers
- Use the Nucleotide collection (nt/nt) database in BLAST13 to analyze each primer.
- To perform the first BLAST analysis, select the Refseq_rna Data Base and filter the search with the group of genera that belong to the subfamily Orthocoronavirinae. They are Alphacoronavirus (taxid:693996), Gammacoronavirus (taxid:694013), and Deltacoronavirus (taxid:1159901). Additionally, evaluate the sequence against other viruses that are co-circulating as H1N1 subtype (taxid:114727), Influenza A virus (taxid:11320), and Influenza B virus (taxid:11520).
- To perform the second BLAST analysis, select the Betacoronavirus GenBank and filter the search with Coronaviridae (taxid:11118) and SARS (taxid:694009). These groups contain sequences of all identified SARS Coronavirus genomes, including genomes found in bats, Betacoronavirus (taxid:694002).
- For this protocol, ensure that the primers do not align with genomes other than the target genome, SARS-CoV-2.
- Primer preparation
- Spin the vials containing the Iyophilized primers with a microcentrifuge (10,000 x g, 1 min at room temperature [RT]) to avoid losses during tube opening.
- Rehydrate the Iyophilized powder in 0.1% diethyl pyrocarbonate (DEPC) water or nuclease-free water to a final concentration of 100 µM (Table 2) and thoroughly dissolve by pipetting up and down. Then, spin at maximum speed (10,000 x g, 1 min at RT) in a microcentrifuge to collect all of the primer solutions at the bottom of the tube.
- Prepare the 10x primer mix under a biosafety cabinet with the forward inner primer (FIP), backward inner primer (BIP), forward outer primer (F3), backward outer primer (B3), loop backward (LB), and loop forward (LF) primers, as reported in Table 2. To prevent losses, pipette or gently vortex the primer solution before performing a rapid spin (10,000 x g, 1 min at RT) with a microcentrifuge.
- Store the 10x primer mix at −20 °C for long-term storage; however, prepare enough for a maximum of five experiments, regardless of several samples to avoid too many freeze-thaw cycles.
NOTE If a smaller volume of the primer mix is needed, then adjust the values by calculating the new volumes (Table 2). Furthermore, the RdRp and RdRp/Hel sets do not include the LF primer because loop primers are not required for RT-LAMP reactions. As a result, replace the volume of LF primer with nuclease-free water or 0.1% DEPC water.
2. RT-LAMP reaction
- Turn on the laminar flow cabinet according to the manufacturer's instructions and wait for at least 3 min for the airflow to stabilize.
- Once the airflow is stable, clean and sanitize the internal surfaces of the cabinet using an aseptic technique. To accomplish this, use the following disinfectants in this order: 1000 ppm quaternary ammonium (benzalkonium chloride), 2% hypochlorite, 3% hydrogen peroxide, and 70% ethanol.
NOTE: In this case, the aseptic technique entails applying the disinfectant and removing it with napkins from inside the cabin to the outside without going over previously cleaned surfaces. - Using the disinfectants from step 2.2, clean the materials that will enter the cabin in the same order.
NOTE: Micropipettes, filter tip boxes, flasks with 1.5 mL and 0.6 mL tubes, 0.2 mL PCR tubes, racks, and a 400 mL beaker must be brought into the cabinet. - Bring some napkins and nitrile gloves into the cabin. After that, turn off the cabinet and expose it to ultraviolet (UV) light for 15 min.
CAUTION: To avoid tissue and DNA damage from prolonged radiation exposure, avoid UV light until the time set in step 2.4 expires.
NOTE Perform the assembly shown in Figure 2 before beginning the protocol, and begin the water bath after completing step 2.4. It is crucial to fill the metal container almost to the brim with drinking water and set the temperature of the iron laboratory heating plate to 90 °C, as this will result in a temperature of ~66.3 °C in the system, which is monitored with the mercury thermometer. - After the irradiation period has ended, restart the cabinet and follow the recommendations in step 1.1.
- Place the reagents (Table 3, Table 4, and Table 5) in an ice-filled cooler or small polystyrene refrigerator. Put the container within the cabinet after cleaning it with 70% ethanol.
- In a 0.6 mL microcentrifuge tube, prepare the LAMP mix of the gene to be amplified (RdRp, N-A, and RdRp/Hel), adding only the following components: 10x Buffer, MgSO4, dNTPs, 1x primers mix and nuclease-free water or 0.1% DEPC water; mix well by pipetting to homogenize.
CAUTION: Because of improper handling and behavior inside the cabinet, there is a high risk of reagent contamination. The following rules must be followed to mitigate this problem: (i) use sterile and filter tips; (ii) use one tip for each reagent; (iii) move slowly and carefully to avoid disrupting the laminar flow; (iv) keep order and use the fewest materials; and (v) use different gloves to prepare the mix and add the genetic material.
NOTE: Keep all the reagents, especially enzymes, on ice because temperature changes can denature them and alter polymerase activity. - Place the 0.6 mL tube(s) with the cap closed in a heating block and incubate at 95 °C for 5 min.
NOTE: Turn on the heating block for 1.5-2.0 mL tubes located outside the cabinet for at least 30 min before beginning the LAMP mix preparation and monitor the temperature (95 °C) with a mercury or alcohol thermometer. - When the incubation is complete, place the tubes in an ice-filled polystyrene cooler for 5 min.
- Return the tubes to the laminar flow cabinet and complete the LAMP mix preparation by adding the enzymes DNA polymerase (Bst 3.0), reverse transcriptase, and high-fidelity DNA polymerase (Table 3, Table 4, and Table 5). In the case of using colorimetric detection, add the dye hydroxinaphthol blue (HNB).
- After adding these reagents, mix the LAMP reagents very well by pipetting them to solubilize the enzymes and dye.
- Fill each PCR tube with 22.0 µL of the mix, being careful not to create bubbles. Then, add 3.0 µL of 0.1% DEPC water or nuclease-free water to the negative control or tube no template control (NTC) and set aside the remaining tube(s) for the addition (genetic material).
NOTE : Keep the PCR tubes in an ice-filled cooler until the sample is added to avoid activating the Bst 3.0 enzyme and starting the reaction prematurely. - Remove all materials from the cabinet and use 70% ethanol to clean the surfaces. Then turn it off following the manufacturer's instructions.
- In a separate area, add 3 µL of the sample to each PCR tube and thoroughly homogenize it. Use a 20 µL micropipette and filter tips to accomplish this.
CAUTION: The micropipette used to add the genetic material must be used exclusively for this purpose and cannot be used to prepare the mix. In this way, contamination of the reagents is avoided. Additionally, keep RNA samples on the ice at all times to reduce the possibility of RNA degradation. Use the following personal protective equipment (PPE) for the sample addition: disposable gown, cap, N95 mask, leggings, lab goggles, and nitrile gloves. - Before performing the colorimetric reaction, take photographs of the PCR tubes with a high-quality camera. The starting color with HNB is violet.
- Carry out the reaction in the following system or equipment: (i) thermal cycler and (ii) water bath.
- Thermal cycler: Deposit the tubes into the reaction block and set up the thermoprofile (see Table 6) on the equipment.
- Water Bath: Deposit the tubes in circular containers and adjust them very well to prevent them from coming out. After that, place the containers in the water bath (Figure 2A, B) at the temperature listed in Table 6.
- In the case of the water bath, once the tubes are inside the system, start the timer for 60 min (Table 6).
- Remove the tubes from the thermal cycler or water bath after the reaction time and store them at 4 °C for the electrophoretic run or at −20 °C until use.
- If a colorimetric reaction was performed, take photographs of the PCR tubes using a high-quality camera. The final color with HNB is sky blue.
3. Analysis of amplification products in agarose gel
NOTE: These steps are suggested as additional checks to colorimetric reaction or control for performance during the standardization step. This is because the technique could present a huge contamination risk to the lab doing these tests.
- Place the bed inside the electrophoresis chamber so that the edge rubbers touch the walls, creating a sealed space for the addition of agarose (internal chamber) (Figure 3A, B).
- After completing step 3.1, weigh the necessary amount of agarose in a 500 mL beaker to obtain a 2% gel. After that, add the required volume of 0.5x Tris-acetate EDTA (TAE) buffer and microwave for 1-2 min.
NOTE: The agarose is completely melted when it is translucent and lump-free when removed from the oven. If this is not confirmed, poorly gelled regions may remain, causing the electrophoretic run and visualization of the amplification products to be altered. - Take the beaker out of the oven and pour the agarose into the internal chamber created in step 3.1 (Figure 3C). Subsequently, check that there are no bubbles, and if there are, remove them using a micropipette tip.
- Arrange the comb to form the wells and leave the agarose to gel for about 30 min at room temperature (RT).
- After this time, add 5 mL of 0.5x TAE buffer to facilitate the removal of the combs and the bed containing the gel. Then position the gel in such a way that the wells are in the anode (Figure 3D).
- Fill the electrophoresis chamber with 0.5x TAE buffer to the capacity specified by the manufacturer, ensuring that the electrodes are in contact with the buffer.
- Add 3 µL of molecular weight marker to the first well of the gel and add 9 µL of NTC and each sample to the following wells. Make these by combining 7 µL of the amplification product with 3 µL of loading buffer; then load 9 µL of this mixture into the gel's wells.
- Cover the electrophoresis chamber with the lid and connect the cables to the power supply ports in the color pattern. Set the power source to the following parameters: 100 V and constant amperage for 120 min.
- After the electrophoretic run is completed, place the gel in the container with the staining solution (ethidium bromide) and incubate for 30 min.
- After incubation, remove the gel from the staining solution and place it in a zip-lock bag. This prevents contamination of the equipment that will be used to visualize the amplicons.
- Visualize the gel on a transilluminator or imager like the Amersham Imager 600.
Results
The implementation of the protocol starts by designing the set of primers for each target gene following the protocol described above. In June 2020, 5,000 SARS-CoV-2 genomes were obtained from the NextStrain database, with a 10% representativeness of Colombian genomes. These sequences were aligned to obtain the consensus sequence that was used in the primer design process. Table 1 shows the primers set chosen for primers RdRp/Hel and RdRp. The primer set for gene N amplification was obtained from a previ...
Discussion
Although the RT-LAMP is regarded as a complementary methodology for performing molecular diagnostics, it also has some limitations and critical steps that must be considered when the protocol is standardized and implemented.
The LAMP standardization for the detection of SARS-CoV-2 evaluated the following parameters and components in the master mix: (a) concentration and temperature of alignment of the primers; (b) concentration of the enzymes; (c) magnesium concentration; (d) reaction time; (e...
Disclosures
Natalia Campillo-Pedroza is CEO of the company BioDx: Diagnóstico y Soluciones Biotecnológicas S.A.S. The rest of the authors declare no conflict of interest.
Acknowledgements
This work was funded by Sistema General de Regalías of Colombia, grant number BPIN 2020000100092, and Universidad Icesi - Convocatoria Interna, grant number CA0413119. MFVT was also financed by the Assistant Professorship Funds from Universidad de los Andes. The funding entities did not participate in the design, execution of activities, data collection, and data analysis and preparation of the manuscript. We thank to University Hospital Fundación Valle del Lili for viral RNA from Sars -CoV-2 samples and Dr. Alvaro Barrera-Ocampo for the comments on the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| 1 kb DNA Ladder | SOLIS BIODYNE | 07-12-00050 | Store at -20 °C |
| 50x TAE Electrophoresis Buffer | ThermoScientific | B49 | Store at roome temperature |
| Accuris High Fidelity Polymerase | ACCURIS LIFE SCIENCE REAGENTS | PR1000-HF-200 | It can be used in case Q5 High-Fidelity DNA polymerase cannot be purchased. For the enzyme, make aliquots of an appropriate volume and store at -20 °C until use. |
| Agarose | PanReacAppliChem | A8963,0100 | N/A |
| Bst 3.0 DNA Polymerase 8000 IU/mL | New England BioLabs | M0374S/M0374L | For the enzyme, make aliquots of an appropriate volume and store at -20 °C until use. |
| Deoxynucleotide (dNTP) Solution Set | New England BioLabs | N0446S | Store at -20 °C |
| Diethyl pyrocarbonate | Sigma-Aldrich | 159220-25G | Handle it with caution under an extraction cabinet |
| GeneRuler 100 bp Plus DNA Ladder, ready-to-use | ThermoScientific | SM0322 | Store at -20 °C |
| Hydroxy naphthol blue disodium salt | Santa Cruz Biotechnology | sc-215156B | N/A |
| Q5 High-Fidelity DNA polymerase 2000 IU/mL | New England BioLabs | M0491S/M0491L | For the enzyme, make aliquots of an appropriate volume and store at -20 °C until use. |
| WarmStart RTx Reverse Transcriptase 15000 IU/mL | New England BioLabs | M0380S/M0380L | For the enzyme, make aliquots of an appropriate volume and store at -20 °C until use. |
References
- World Health Organization. . Who coronavirus (COVID-19) dashboard (no date). , (2023).
- Ibrahim, N. K. Epidemiologic surveillance for controlling Covid-19 pandemic: types, challenges and implications. Journal of Infection and Public Health. 13 (11), 1630-1638 (2020).
- Rojas-Gallardo, D. M., et al. COVID-19 in Latin America: Contrasting phylodynamic inference with epidemiological surveillance. (Molecular epidemiology of COVID-19 in Latin America). medRxiv. , (2020).
- Liu, R., et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clinica Chimica Acta. 505, 172-175 (2020).
- Kevadiya, B. D., et al. Diagnostics for SARS-CoV-2 infections. Nature Materials. 20 (5), 593-605 (2021).
- Tomita, N., Mori, Y., Kanda, H., Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nature Protocols. 3 (5), 877-882 (2008).
- Li, Y., Fan, P., Zhou, S., Zhang, L. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microbial Pathogenesis. 107, 54-61 (2017).
- Notomi, T., Mori, Y., Tomita, N., Kanda, H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. Journal of Microbiology. 53 (1), 1-5 (2015).
- Augustine, R., et al. Loop-mediated isothermal amplification (LAMP): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology (Basel). 9 (8), 182 (2020).
- . Nextstrain Available from: https://nextstrain.org/ (2023)
- . Neb Lamp, NEB LAMP Available from: https://lamp.neb.com/ (2023)
- . Blast: Basic local alignment search tool (no date) Available from: https://blast.ncbi.nlm.nih.gov/ (2023)
- Zhang, Y., et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. , (2020).
- Lu, R., et al. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virologica Sinica. 35 (3), 344-347 (2020).
- Najafov, A., Hoxhaj, G. . PCR Guru. , (2017).
- Zhang, Y., et al. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques. 69 (3), 178-185 (2020).
- Ramírez-Chavarría, R. G., et al. Automatic analysis of isothermal amplification via impedance time-constant-domain spectroscopy: A SARS-CoV-2 case study. Chemosensors. 11 (4), 230 (2023).
- Haque, M. F. U., et al. A novel RdRp-based colorimetric RT-LAMP assay for rapid and sensitive detection of SARS-CoV-2 in clinical and sewage samples from Pakistan. Virus Research. 302, 198484 (2021).
- Donia, A., et al. Integration of RT-LAMP and microfluidic technology for detection of SARS-CoV-2 in wastewater as an advanced point-of-care platform. Food and Environmental Virology. 14, 364-373 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved