A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
A Clinical Metaproteomics Workflow Implemented within Galaxy Bioinformatics Platform to Analyze Host-Microbiome Interactions Underlying Human Disease
In This Article
Summary
Clinical metaproteomics offers insights into the human microbiome and its contributions to disease. We harnessed the computational power of the Galaxy platform to develop a modular bioinformatics workflow that facilitates complex mass spectrometry-based metaproteomic analysis and characterization of diverse clinical sample types relevant to studies of disease.
Abstract
Clinical metaproteomics reveals host-microbiome interactions underlying diseases. However, challenges to this approach exist. In particular, the characterization of microbial proteins present in low abundance relative to host proteins is difficult. Other significant challenges are attributed to using very large protein sequence databases, which impedes sensitivity and accuracy during peptide and protein identification from mass spectrometry data in addition to retrieving taxonomy and functional annotations and performing statistical analysis. To address these problems, we present an integrated bioinformatics workflow for mass spectrometry-based metaproteomics that combines custom protein sequence database generation, peptide-spectrum match generation and verification, quantification, taxonomic and functional annotations, and statistical analysis. This workflow also offers characterization of human proteins (while prioritizing microbial proteins), thus offering insights into host-microbe dynamics in disease. The tools and workflow are deployed in the Galaxy ecosystem, enabling the development, optimization, and dissemination of these computational resources. We have applied this workflow for metaproteomic analysis of numerous clinical sample types, such as nasopharyngeal swabs and bronchoalveolar lavage fluid. Here, we demonstrate its utility via the analysis of residual fluid from cervical swabs. The complete workflow and accompanying training resources are accessible on the Galaxy Training Network to equip non-experts and experienced researchers with the necessary knowledge and tools to analyze their data.
Introduction
Mass spectrometry (MS)-based metaproteomics identifies and quantifies microbial and human proteins from clinical samples. This approach provides a new understanding of microbiome responses to disease and uncovers potential mediators of host-microbiome interactions1,2. Although metaproteomic analysis of clinical samples can uncover the microbiome's interactions with its host environment, the field still faces many challenges. One main challenge is the relatively high abundance of host (human) proteins, which hampers the identification of lower abundant microbial proteins. Moreover, MS-based metaproteomics depends on the use of very large protein sequence databases. These databases comprise microbial proteomes that are present in the sample, which can result in a large database containing millions of sequences. Following the generation of tandem mass spectrometry (MS/MS) spectra from tryptically digested proteins, the MS/MS spectra are searched against large protein sequence databases, matching a peptide sequence to each spectrum (peptide-spectrum match, or PSM). However, sensitivity decreases, and the potential for false positives increases with large databases used for metaproteomics3. Additionally, conserved protein sequences across taxa and insufficient annotation of encoded proteins limit taxonomic and functional annotations for detected peptides and proteins4,5. We present a bioinformatics workflow for effective metaproteomic analysis of clinical samples that addresses many of these challenges and provides accessible software resources for researchers to investigate host-microbiome dynamics underlying human disease.
Clinical metaproteomics has been used to investigate diverse sample types, including feces and vaginal swabs, among others, to decipher pathogenic mechanisms in diseases and conditions6,7,8,9,10,11,12,13,14,15,16,17,18,19,20. Here, we use a metaproteomic bioinformatics workflow to analyze a subset of MS/MS data from Pap test fluid (PTF) samples from ovarian cancer (OVCA) and non-OVCA patients21. The software tools and workflow are accessible via the Galaxy platform, which streamlines the development and execution of complex clinical metaproteomic workflows22,23,24,25. Galaxy is an open-source platform that is designed for bioinformatics and computational biology. It provides a web-based environment for the use of open-source tools and workflows where academic researchers can perform and share complex data analyses. A thriving global community of software developers, data scientists, and end-users maintains the Galaxy ecosystem, including the Galaxy Training Network (GTN; https://training.galaxyproject.org/), which offers online and on-demand training resources22,23,24,25,26,27. Our workflow aims to reveal a new understanding of host-microbe dynamics in clinical samples as well as generate novel, well-characterized peptide targets of interest for developing targeted MS-based clinical assays for further study of clinical samples6,20,28. Furthermore, this manuscript intends to highlight the clinical metaproteomics workflow methodology. More detailed and beginner-friendly guides are provided in the GTN (https://training.galaxyproject.org/) as it is a valuable resource that can be used in parallel with this manuscript for users seeking additional explanations not covered. The Galaxy community has authored numerous manuscripts to aid beginner users of the Galaxy platform20,21,22,23,24,25,26,27.
All supplementary tables (e.g., tool parameters) and figures (e.g., example plots) for this manuscript have been provided as separate files and are referenced accordingly. Current tool versions within Galaxy version 2.3.0 were used for this manuscript. Therefore, results may differ slightly depending on Galaxy and tool version updates. The Galaxy platform and its tools are open-source and can be used for academic research purposes.
Access restricted. Please log in or start a trial to view this content.
Protocol
MS/MS spectral data were obtained from de-identified residual PTF samples that were collected using procedures that followed institutional board-approved guidelines and regulations, as previously described21,29,30.
NOTE: Figure 1 provides an overview of the complete workflow, which consists of five modules. All inputs, outputs, and software tools are summarized in Supplementary Table 1.
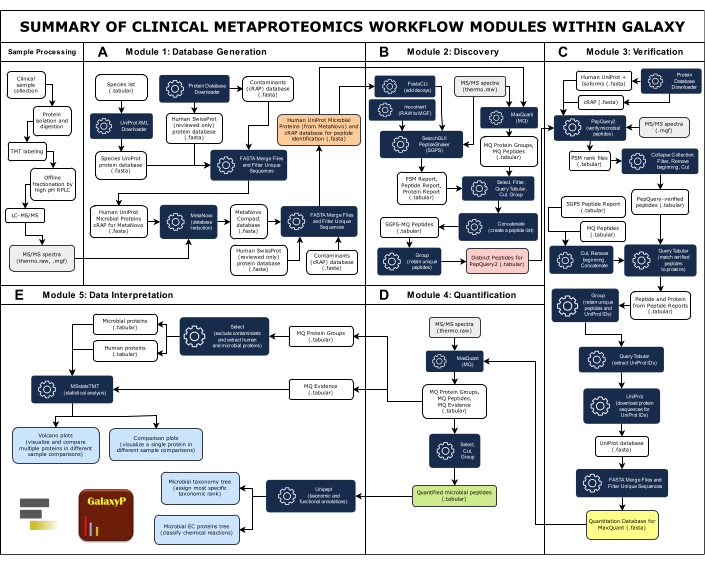
Figure 1: Summary of Clinical Metaproteomics Workflow Modules Within Galaxy. The complete clinical metaproteomics workflow comprises five modules: Database Generation, Discovery, Verification, Quantification, and Data Interpretation. (A) The large comprehensive database includes protein sequences from microbial species thought to be present in the sample, humans, and common contaminants. The MetaNovo software tool directly matched MS/MS spectral data to peptides and infers proteins and their source organism from raw MS data and the large input protein sequence database, creating a reduced database33. The reduced database from MetaNovo is then merged with human and contaminant proteins to create the database for peptide discovery. (B)Two peptide identification algorithms, SearchGUI/PeptideShaker, and MaxQuant, match peptide sequences to MS/MS spectra and the target-decoy protein database49. (C)Peptides identified by SearchGUI/PeptideShaker and MaxQuant are next verified using PepQuery2. PepQuery2 rigorously re-examines putatively identified microbial peptide sequences and their matched MS/MS spectra against other potential matches to the human host proteome and/or contaminants, thereby verifying high-confident microbial matches40,41. Verified peptides are used to generate a verified protein sequence database that will be used for peptide and protein quantification. (D) MaxQuant42 searches MS/MS data against the verified protein sequence and quantifies microbial peptides and inferred proteins along with human proteins. (E) Unipept45 and MSstatsTMT46 are used in the final step to annotate proteins with taxonomy and functional information (enzyme commission accessions) as well as generate volcano and comparison plots. Please click here to view a larger version of this figure.
1. TMT labeling and generation of MS/MS spectra
- To prepare for MS analysis, perform clinical sample collection per guidelines and regulations.
NOTE: Because this protocol emphasizes the bioinformatic workflow, procedures for clinical sample collection may differ from what was used for this manuscript. Here, proteins were tryptically digested into a peptide mixture, labeled, fractionated, and analyzed via mass spectrometry to generate MS/MS spectral data for downstream analysis using the Galaxy platform. Detailed sample processing instructions have been previously described by Boylan et al.29 and Afiuni-Zadel et al.30. - Isolate proteins from clinical samples and digest them into peptides using trypsin29,30.
- Label proteins with a Tandem Mass Tag (TMT)-11-plex reagent. This tagging reagent will aid in quantifying peptides and proteins31,32.
- Divide labeled samples randomly and evenly into four TMT-based experimental groups.
- For each experimental group, include one pooled reference sample labeled with a unique TMT tag to serve as the common reference for comparison to each individual sample across the four experimental groups31,32.
- Perform offline fractionation on pooled samples by high pH reversed-phase liquid chromatography (RPLC)29,30.
- Analyze the fractions by liquid chromatography-tandem MS (LC-MS/MS) via a hybrid quadrupole-Orbitrap mass spectrometer29,30. Save the generated MS/MS spectral data in Thermo Raw format (thermo.raw).
NOTE: As required, Thermo Raw files are converted to Mascot Generic Format (.mgf) to be compatible with various software. In this text, the abbreviations "RAW" and "MGF" denote the file format of the input MS/MS data sets. In the figures, the MS/MS data sets are represented by the same RAW icons for simplicity.
2. Module set up
NOTE: Button/menu selections are bolded. Example files, workflows, and tool parameters are accessible via Supplementary tables. More information on how to use Galaxy can be found on the GTN FAQs page (https://training.galaxyproject.org/training-material/faqs/galaxy/).
- Galaxy Europe server
- Access the Galaxy Europe server (Galaxy EU; https://usegalaxy.eu/).
- Create an account or login. A valid email address is required to create a new account. Log in as a user to use Galaxy.
- Preparing a Galaxy history
- If a user is importing example inputs from Supplementary Table 2 follow steps 2.2.1.1-2.2.1.3.
- Open the example Galaxy histories using the links provided in Supplementary Table 2.
- Click the gray Import this history button located in the top-left corner of the (center) panel. Rename the history and click Copy History. If desired, add their data sets to this history by clicking the Upload button in the far-left panel and add files for upload.
- Click Start > Close. The uploaded file(s) will appear in the history panel on the right-hand side. Wait for the color of the data set(s) to turn green before using.
NOTE: If importing (copying) an existing history, do not create a separate (new) history.
- If a user is creating a new history and uploading their data, follow steps 2.2.2.1.-2.2.2.2.
- On the History panel (right side), click + (plus) icon once to create a new history called "Unnamed History". Click the pencil icon next to the history and click Save. The same steps for adding data sets to an existing (example) history apply to uploading one's data.
- In the far-left panel, click Upload and add files for upload. Click Start > Close. The uploaded file(s) will appear in the new history. Wait for the color of the data set(s) to turn green.
- If a user is analyzing multiple MS/MS files simultaneously follow steps 2.2.3.1.-2.2.3.3.
- Place them into a data set collection to select them as one input. Click the check mark icon on the History panel and select (check) data sets.
- Click on the button that says the number of selected data sets (e.g., 4 of 8 selected), and in the drop-down menu, click Build Dataset List. In the pop-up window, type a name for the collection (e.g., MGF Data, RAW Data). If desired, select if the original data sets will be hidden once the collection is made.
- Click the blue Create Collection button in the bottom-right corner of the pop-up. Click on the check mark icon in the History panel to deselect the data sets.
NOTE: Each of the five modules should be run in its own (imported or new) Galaxy history for improved user experience. To avoid redundancy, later module instructions will omit set-up and focus on workflow steps.
- If a user is importing example inputs from Supplementary Table 2 follow steps 2.2.1.1-2.2.1.3.
- Importing and running a workflow
NOTE: It is strongly advised for all users, whether using example data or their data, to use and/or adapt the modular workflows with preset parameters (Supplementary Table 2). In doing so, users can avoid having to search for and set the parameters for each tool. If desired, users can search for tools by clicking on the Tools button in the far-left panel and type in the tool name (as accurately as possible) into the search bar in the adjacent panel. Matching tools will pop up automatically. Click on the correct search result and set the appropriate parameters (refer to Supplemental File 1). Before running a tool, users can set up email notifications to alert them when a job has been completed by selecting the button near the end of the parameters. For convenience, there are two Run buttons: one at the top-right corner of the center panel and the other after the parameter fields. Supplementary Table 3 provides additional training resources. Tool versions and databases are current and operational at the time of writing (June 2024) but may change as Galaxy and associated tools and databases are updated.- Open the workflow in a new tab using the links in Supplementary Table 2.
- Click the Import button in the top-right corner of the panel. A new tab will open with a green box that confirms that the workflow has been imported. The green box will also include two options: start using this workflow right away or return to the previous page.
- Click the first button ("start using this workflow…") to open the Workflow tab in the center panel of the interface, which displays all stored workflows. Find the workflow that was just imported and click the blue play (triangle) button. This will display the input fields.
NOTE: For each provided workflow, the input fields correspond to the example inputs (Supplementary Table 2). If a user is analyzing their data, their inputs should be named accordingly to ensure the correct files are used for each module.
- If a user wants to view workflows on the Galaxy EU server, follow steps 2.3.2.1-2.3.2.4.
- Click the Workflow button in the top bar of the Galaxy website. Within this tab, click the sub-tab My workflows to display all imported workflows. To view a workflow, click the Edit button that has a pencil icon to open the Workflow Editor.
- Within the Workflow Editor, interact with the workflow, such as clicking and dragging to re-organize, clicking on the tools to view them, changing parameters, etc. After making changes, save the edited workflow by clicking on the disk icon at the top of the right panel, and if desired, run the workflow by clicking the play icon (also at the top of the right panel).
- Create user-specific workflows to analyze custom input data. Depending on the user's knowledge of metaproteomics and experience with the Galaxy platform, build a workflow and then analyze the data.
- If a user is less experienced, test various tools in history and then extract a workflow from their completed analysis.
NOTE: This extracted workflow can be expanded, revised, and reused, allowing users to reproduce their work accurately. More detailed instructions can be found at the GTN FAQs section for workflows (https://training.galaxyproject.org/training-material/faqs/galaxy/#workflows).
- Click on each input field and select the appropriate input. Sections 3 through 7 describe module inputs. Check that all inputs are in an accepted format to avoid errors. Click accepted formats under each input field to check if all files are compatible with the tools. Once done, click Run workflow.
NOTE: If a user prefers to set up the tools manually, tutorial material for each module of this clinical metaproteomics workflow is provided on the GTN website (https://gxy.io/GTN:P00019). Estimated runtimes for key tools have been included in Supplementary Table 2, but runtimes are dependent on input data size, tool dependencies (such as memory requirements compared to allocated memory), scheduled maintenance times, errors, etc. Job statuses are indicated by the color of the data set, and when the data set is selected (clicked), a message will appear that states whether a job is waiting to be queued (gray), running (orange), or failed (red). When a job has completed, the data set will turn green (no confirmation message). Users can opt-in to email notifications to alert them when jobs have finished (see NOTE at the beginning of step 2.3). Module instructions below will omit explicit set-up steps as they are the same for each module (see section 2 and GTN FAQs if needed) and will describe the key tools for each module. See Supplementary Table 1 for a complete list of tools used. Tool names have been bolded. For reference, all tool names, versions, and descriptions are included in the Table of Materials. If a user is running the example workflows from Supplementary Table 2, refer to the example file names included in the parentheses at the end of each step. If a user is running the tools independently, the example file names can be disregarded. To rename a data set, click on the pencil icon in the top-right corner of the data set. In the "Name" field, type the new name, and click Save.
- Open the workflow in a new tab using the links in Supplementary Table 2.
3. Module 1: Protein sequence database generation
NOTE: If a user wants to use the example inputs and workflow from Supplementary Table 2, be sure to follow the instructions in section 2. For Module 1, import the input and workflow for DATABASE GENERATION. The output column of Supplementary Table 2 includes examples of completed output histories for reference. For all modules, the corresponding GTN tutorial can be found in Supplementary Table 3.
- Compile a list of species that are linked to the disease or condition of interest and/or the site of sample collection.
- Obtain this species list from a literature review. Alternatively, if the samples have been previously analyzed, obtain the species list from 16S rRNA or metagenomic sequencing.
- Save this species list as a tabular file (e.g., Species.tabular).
NOTE: Using the species list, a large comprehensive database of protein sequences of known disease-causing microorganisms will be generated, and using MetaNovo, this large database, which contains millions of protein sequences, will then be reduced to a more manageable database that contains proteins present in samples. The database reduction step is crucial as many database searching tools cannot handle millions of sequences. The reduced database will be merged with human and contaminant proteins to generate a compact database to generate a compact database, which will be used for peptide identification in the next module (section 4).
- Use the species list (Species.tabular) as input for UniProt (download proteome as fasta) to generate a protein sequence database (Species UniProt FASTA.fasta).
- Run Protein Database Downloader to generate two more protein sequence databases: Human SwissProt (reviewed-only) and contaminant proteins (Human SwissProt Protein Database.fasta, Contaminants [cRAP] Protein Database.fasta). Contaminant proteins are also termed as common Repository of Adventitious Proteins, or cRAP.
- Use the three protein databases as inputs for FASTA Merge Files and Filter Unique Sequences to exclude duplicates and generate a large protein sequence database (Human UniProt Microbial Proteins cRAP for MetaNovo.fasta).
- Use the large (comprehensive) database (from step 3.4), and MS data sets (MGF) as input for MetaNovo33 to generate a reduced database (MetaNovo Compact Database.fasta).
- Run FASTA Merge Files and Filter Unique Sequences on the MetaNovo-generated database, the Human SwissProt (reviewed-only), and cRAP databases to generate a reduced (target) database of microbial, human, and contaminant protein sequences that will be used for detecting peptides (Human UniProt Microbial Proteins [from MetaNovo] and cRAP.fasta).
4. Module 2: Peptide discovery via database searching
NOTE: If a user wants to use the example inputs and workflow from Supplementary Table 2, be sure to follow the instructions in Section 2. For Module 2, import the input and workflow for DISCOVERY. For all modules, the corresponding GTN tutorial can be found in Supplementary Table 3. SearchGUI34,35,36 and PeptideShaker37 are separate software but will be considered as one peptide identification and processing program as they are used in tandem. For software compatibility, the MS/MS data sets will be converted from RAW to MGF for SearchGUI/PeptideShaker using the msconvert tool (in the provided workflow). MaxQuant38 can process RAW files.
- Run FastaCLI to add decoy protein sequences to the reduced (target) database to generate a target-decoy protein sequence database (FastaCLI MetaNovo Human SwissProt cRAP with decoys.fasta).
NOTE: FastCLI will only need to be run for SearchGUI/PeptideShaker. MaxQuant can add decoys and contaminants to a protein sequence database. Here, the reduced database already contains contaminants (cRAP), so MaxQuant has been set to add only decoys. - Run SearchGUI/PeptideShaker and MaxQuant to search the MS data sets against the reduced database to identify peptides and eventually assign them to protein sequences via sequence database searching. See Supplementary Table 4 for tool parameters.
NOTE: Two peptide identification programs will be used here (SearchGUI/PeptideShaker and MaxQuant) to identify peptide and protein sequences via sequence database searching. These programs identify peptides in the MS/MS spectra and search a protein sequence database, matching observed and theoretical peptide data, including peptide masses and spectra. In the following module, identified peptides will be verified using PepQuery2 to validate that microbial peptides were obtained (section 5).- Run SearchGUI to generate an archive file that contains PSMs (Search GUI on data [#].searchgui_archive).
- Use the SearchGUI archive file as input for PeptideShaker to generate a PSM Report, Peptide Report, and Protein Report (Peptide Shaker on data [#]: [report name].tabular).
- Run MaxQuant to generate Protein Groups and Peptides files (MaxQuant Protein Groups.tabular, MaxQuant Peptides.tabular).
NOTE: MaxQuant requires an experimental design file, which contains experimental conditions, sample groups, and relationships between samples (Experimental Design Discovery MaxQuant.tabular). This file informs MaxQuant how to organize and analyze the MS data. An example has been provided in Supplementary Table 5. If using the user's data, users must modify this file to match their MS data sets.
- Use text manipulation tools to manage outputs from both programs. View the DISCOVERY workflow in Supplementary Table 2 to see which tools are applicable for SearchGUI/PeptideShaker and MaxQuant.
NOTE: The following text manipulation tools are implemented in Galaxy. The key tools are highlighted below, so it is highly recommended that users refer to the DISCOVERY Workflow to see additional tools that are not covered here. See section 2 for instructions on how to view a workflow.- Select microbial matches (Select microbial PSMs.tabular from SGPS, Select microbial peptides (MQ).tabular).
- Use Filter and Query Tabular39 to select confident PSMs and query for their protein accession numbers (Filter confident microbial PSMs.tabular, query results on data [# and #].tabular).
- Use Cut to extract peptide sequences as a new data set (Cut on data [#].tabular).
- Use Group to obtain unique entries (e.g., unique peptide sequences) for each program (MQ Peptides.tabular, SGPS Distinct Peptides.tabular).
- Concatenate the two peptide lists into a single data set (SGPS-MQ Peptides.tabular).
- Group to remove duplicate peptide sequences. The final list of distinct microbial peptides will be used for PepQuery2 verification (Distinct Peptides.tabular).
5. Module 3: Verification of microbial peptides
NOTE: If a user wants to use the example inputs and workflow from Supplementary Table 2, be sure to follow the instructions in Section 2. For Module 2, import the input and workflow for VERIFICATION. For all modules, the corresponding GTN tutorial can be found in Supplementary Table 3.
- Use the following as inputs for PepQuery240,41 List of distinct microbial peptides (Distinct Peptides for PepQuery.tabular); MS spectral data sets (MGF); the Human UniProt Reference (along with isoforms) (Human UniProt+Isoforms FASTA.fasta) and cRAP protein sequence databases (cRAP.fasta). See the parameters in Supplementary Table 6.
NOTE: Verifying the presence of peptides and proteins is crucial in obtaining accurate data and significant insights into the proteome of a biological system. PepQuery2 enables the validation of novel, disease-specific peptides of interest with sensitivity and specificity. The identified microbial peptides (from module 2) will be searched against human and contaminant protein sequences to verify that they are of microbial origin (avoid misassignment of human peptides). The verified peptides will be used to generate a sequence database of verified proteins, which is necessary to reduce the introduction of false positives during protein quantification in the following module (section 6).- One PSM rank file will be generated for every MS/MS data set used as input (PepQuery2 on collection [#]: psm_rank.tabular). Run Collapse Collection on the PSM rank files to create one combined data set (Collapse Collection on data [#] .tabular) and Filter to retain confident PSMs (Filter on [PSM rank collection].tabular).
- Run Remove beginning to exclude column headers and Cut to extract the verified peptide sequences as a new data set.
- Run Cut on the Peptide Reports from SearchGUI/PeptideShaker and MaxQuant (SGPS Peptide Report.tabular, MaxQuant Peptide Report.tabular) to extract the peptide sequences and protein entries as a new peptide-protein data set (for each program) and Remove beginning to exclude the column headers.
- Concatenate the peptide sequences and protein entries from both programs to create a new (combined) peptide-protein data set.
- Run Query Tabular on the combined peptide-protein data set and the verified peptides to assign the verified peptides to their associated protein entries (Peptide and Protein from Peptide Reports.tabular). Protein entries are cataloged by their protein accession numbers (also known as UniProt IDs).
- Group to retain unique verified peptides and their associated UniProt IDs.
- Run Query Tabular to extract the UniProt IDs (UniProt-ID from verified Peptides.tabular).
- Put the UniProt IDs into UniProt to obtain their associated protein sequences as a new database (UniProt.fasta).
- Run FASTA Merge Files and Filter Unique Sequences on the UniProt-generated protein sequence database, the Human UniProt database(alongwith isoforms), and contaminant databases to generate a verified database that will be used for peptide quantification (Quantitation Database for MaxQuant.fasta).
6. Module 4: MaxQuant quantification
NOTE: If a user wants to use the example inputs and workflow from Supplementary Table 2, be sure to follow the instructions in Section 2. For Module 2, import the input and workflow for QUANTIFICATION. For all modules, the corresponding GTN tutorial can be found in Supplementary Table 3.
- Use the verified protein sequence database and MS data sets (RAW) as inputs for MaxQuant42.
NOTE: Remember that MaxQuant requires an experimental design file and can be the same file as the one used for peptide identification (step 4.2). Change file names as needed. The verified database from the previous module is required to reduce false positives during protein quantification. Protein quantification enables researchers to measure and compare peptide and protein abundances in biological samples. This step is imperative to understanding differential protein expression by obtaining insights into quantitative changes across different conditions.- Generate the Evidence, Protein Groups, and Peptides files (MaxQuant Evidence.tabular, MaxQuant Protein Groups.tabular, MaxQuant Peptides.tabular).
- Select microbial peptides from the MaxQuant Peptides file (Select microbial peptides.tabular).
- Cut out only the microbial peptide sequences (Cut on data [#].tabular).
- Group to obtain a list of quantified microbial peptides (Quantified Peptides.tabular).
7. Module 5: Data interpretation
NOTE: If a user wants to use the example inputs and workflow from Supplementary Table 2, be sure to follow the instructions in Section 2. For Module 2, import the input and workflow for DATA INTERPRETATION. For all modules, the corresponding GTN tutorial can be found in Supplementary Table 3. Outputs from MaxQuant quantification in the previous module will be used here for taxonomic and functional annotations using Unipept and statistical analysis using MSstatsTMT. Unipept enables researchers to identify and quantify microorganisms within diverse environments and integrates with public databases (like UniProt) to retrieve updated annotations. MSstatsTMT was designed for robust statistical analysis of mass spectrometry-based quantitative proteomics data using TMT labeling.
- Use the list of quantified microbial peptides (Quantified Peptides.tabular) as the input for Unipept43,44,45 to perform taxonomic and functional annotations. See Supplementary Table 7 for parameters and a list of outputs.
- Unipept outputs of interest here are the microbial taxonomy tree and a microbial enzyme commission (EC) proteins tree (Microbial Taxonomy Tree.d3_hierarchy, Microbial EC Proteins Tree.d3_hierarchy).
- To view the trees, click on the data set to open the options. Click on Visualize (4th option from the left) > Unipept Taxonomy Viewer.
- To view taxonomic and functional annotations in a table (Unipept peptinfo.tabular): click on the eye icon in the top right corner of the data set. Scroll to see each peptide on its own row and information across different columns.
- Before performing statistical analysis using MSstatsTMT, run Select on the MaxQuant Protein Groups file to create two new data sets: microbial and human proteins (Microbial Proteins.tabular, Human Proteins.tabular). Proteins have taxonomy tags that designate their origin.
- Exclude contaminant proteins with the tag "con_."
- Retain microbial and human proteins, which are designated with microbial (e.g., "_9LACO") and "_HUMAN" tags, respectively (Microbial-Proteins.tabular, Human-Proteins.tabular).
- MSstatsTMT42,46,47 will be used to perform statistical analysis. Use the MaxQuant Evidence file (from Module 4) and the selected microbial proteins (or human proteins) from the previous step as inputs. This workflow prioritizes microbial proteins but offers the option to characterize human proteins as well. See Supplementary Table 8 for parameters and a list of outputs.
NOTE: MSstatsTMT requires an annotation file and a comparison matrix (also known as a contrast matrix). The annotation file will determine how quantifications will be combined, while the comparison matrix will accommodate different sample groups. Examples of these files have been included (Annotation.tabular, Comparison Matrix.tabular) in Supplementary Table 9 and Supplementary Table 10. - MSstatsTMT outputs of interest here are the volcano and comparison plots for the microbial proteins (Microbial Proteins Volcano Plot.pdf, Microbial Proteins Comparison.pdf). View the plots by clicking on the eye icon in the top right corner of the data set.
Access restricted. Please log in or start a trial to view this content.
Results
The general protocol described here was demonstrated on MS/MS files obtained from a subset of PTF samples21. Do et al.21 analyzed four MS/MS files from PTF samples that were collected following procedures described by Boylan et al.29and Afiuni-Zadel et al.30. This workflow prioritizes microbial proteins but offers the flexibility for the characterization of human proteins in parallel with microbial proteins21...
Access restricted. Please log in or start a trial to view this content.
Discussion
Clinical metaproteomics research offers potential breakthroughs for clinical studies, but challenges in its implementation persist. The lower abundance of microbial proteins relative to the host proteins in most samples hinders the detection and characterization of non-host proteins6,10. Dependence on large protein sequence databases for accurate peptide and protein identification and quantification, along with complexities of taxonomically and functionally annot...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors declare no conflict of interest.
Acknowledgements
We thank Dr. Amy Skubitz and Dr. Kristin Boylan (University of Minnesota) for the pilot data sets and Dr. Paul Piehowski, Dr. Tao Liu, and Dr. Karin Rodland (Pacific Northwest National Laboratories (PNNL)) for their expertise in the sample collection, and processing of the PTF samples and generation of the TMT-labeled MS data used in this study. This project was funded in part by the Minnesota Ovarian Cancer Alliance (MOCA), the National Institutes of Health/National Cancer Institute Grant Number: 5R01CA262153 (A.P.N.S.), 1R21CA267707 (P.D.J and T.J.G.), and the National Institutes of Health/National Cancer Institute Grant Number: P30CA077598 (P.D.J. and T.J.G.).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Collapse Collection | GalaxyP | Galaxy Version 5.1.1 | Combines a dataset list collection into a single file (in the order of the list) |
| Concatenate datasets | GalaxyP | Galaxy Version 0.1.1 | Concatenate files tail-to-head |
| Cut | GalaxyP | Galaxy Version 1.0.2 | Cut (select) specified columns from a file |
| FASTA Merge Files and Filter Unique Sequences | GalaxyP | Galaxy Version 1.2.0 | Concatenate FASTA database files together |
| FastaCLI | GalaxyP | Galaxy Version 4.0.41+galaxy1 | Appends decoy sequences to FASTA files |
| FASTA-to-Tablular | GalaxyP | Galaxy Version 1.1.0 | Convert FASTA-formatted sequences to TAB-delimited format |
| Filter | GalaxyP | Galaxy Version 1.1.1 | Filter columns using simple expressions |
| Filter Tabular | GalaxyP | Galaxy Version 3.3.0 | Filter a tabular file via line filters |
| Galaxy Europe (EU) server | GalaxyP | https://usegalaxy.eu/ | |
| Group | GalaxyP | Galaxy Version 2.1.4 | Group a file by a particular column and perform aggregate functions |
| Identification Parameters | GalaxyP | Galaxy Version 4.0.41+galaxy1 | Set identification parameters for SearchGUI/PeptideShaker |
| Learning Pathway: Clinical metaproteomics workflows within Galaxy | GalaxyP | https://training.galaxyproject.org/training-material/learning-pathways/clinical-metaproteomics.html | |
| MaxQuant | GalaxyP | Galaxy Version 2.0.3.0+galaxy0 (Discovery module); Galaxy Version 1.6.17.0+galaxy4 (Quantification module) | Quantitative proteomics software package for analysis of large mass spectrometric data files |
| MetaNovo | GalaxyP | Galaxy Version 1.9.4+galaxy4 | Search MS/MS data against a FASTA database (of known proteins) to produce a targeted database (of matched proteins) for mass spectrometry analysis |
| msconvert | GalaxyP | Galaxy Version 3.0.20287.2 | Convert and/or filter mass spectrometry files |
| MSstatsTMT | GalaxyP | Galaxy Version 2.0.0+galaxy1 | R-based package for detection of differentially abundant proteins in shotgun mass spectrometry-based proteomic experiments using tandem mass tag (TMT) labeling |
| PepQuery2 | GalaxyP | Galaxy Version 2.0.2+galaxy0 | Peptide-centric search engine for identification and/or validating known and novel peptides of interest |
| PeptideShaker | GalaxyP | Galaxy Version 2.0.33+galaxy1 | Interpret results from SearchGUI for protein identification |
| Protein Database Downloader | GalaxyP | Galaxy Version 0.3.4 | Download specified protein sequences as a FASTA file |
| Query Tabular | GalaxyP | Galaxy Version 3.3.0 | Load tabular files intoa SQLite database |
| Remove beginning | GalaxyP | Galaxy Version 1.0.0 | Remove the specified number of (header) lines from a file |
| SearchGUI | GalaxyP | Galaxy Version 4.0.41+galaxy1 | Run search engines on MGF peak lists and prepare results for input to Peptide Shaker |
| Select | GalaxyP | Galaxy Version 1.0.4 | Select lines that match an expression |
| Unipept | GalaxyP | Galaxy Version 4.5.1 | Retrieve UniProt entries and taxonomic information for tryptic peptides |
| UniProt | GalaxyP | Galaxy Version 2.3.0 | Download proteome as a XML (UniProtXML) or FASTA file from UniProtKB |
References
- Zhang, X., Li, L., Butcher, J., Stintzi, A., Figeys, D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome. 7 (1), 154(2019).
- Van Den Bossche, T., et al. The Metaproteomics Initiative: a coordinated approach for propelling the functional characterization of microbiomes. Microbiome. 9 (1), 243(2021).
- Tanca, A., et al. Evaluating the impact of different sequence databases on metaproteome analysis: insights from a lab-assembled microbial mixture. PloS One. 8 (12), e82981(2013).
- Seifert, J., et al. Bioinformatic progress and applications in metaproteogenomics for bridging the gap between genomic sequences and metabolic functions in microbial communities. Proteomics. 13 (18-19), 2786-2804 (2013).
- Muth, T., Renard, B. Y., Martens, L. Metaproteomic data analysis at a glance: advances in computational microbial community proteomics. Expert Rev Proteomics. 13 (8), 757-769 (2016).
- Bihani, S., et al. Metaproteomic analysis of nasopharyngeal swab samples to identify microbial peptides in COVID-19 patients. J Proteome Res. 22 (8), 2608-2619 (2023).
- Ayan, E., DeMirci, H., Serdar, M. A., Palermo, F., Baykal, A. T. Bridging the Gap between Gut Microbiota and Alzheimer's Disease: A metaproteomic approach for biomarker discovery in transgenic mice. Int J Mol Sci. 24 (16), 12819(2023).
- Levi Mortera, S., et al. A metaproteomic-based gut microbiota profiling in children affected by autism spectrum disorders. J Proteomics. 251, 104407(2022).
- Long, S., et al. Metaproteomics characterizes human gut microbiome function in colorectal cancer. NPJ Biofilms Microbiomes. 6 (1), 14(2020).
- Hardouin, P., Chiron, R., Marchandin, H., Armengaud, J., Grenga, L. Metaproteomics to Decipher CF Host-Microbiota interactions: Overview, challenges and future perspectives. Genes (Basel). 12 (6), 892(2021).
- Levi Mortera, S., et al. Functional and taxonomic traits of the gut microbiota in Type 1 diabetes children at the onset: A metaproteomic study. Int J Mol Sci. 23 (24), 15982(2022).
- Gonzalez, C. G., et al. Location-specific signatures of Crohn's disease at a multi-omics scale. Microbiome. 10 (1), 133(2022).
- Thuy-Boun, P. S., et al. Metaproteomics analysis of SARS-CoV-2-infected patient samples reveals presence of potential coinfecting microorganisms. J Proteome Res. 20 (2), 1451-1454 (2021).
- Grenga, L., et al. Taxonomical and functional changes in COVID-19 faecal microbiome could be related to SARS-CoV-2 faecal load. Environ Microbiol. 24 (9), 4299-4316 (2022).
- Biemann, R., et al. Fecal metaproteomics reveals reduced gut inflammation and changed microbial metabolism following lifestyle-induced weight loss. Biomolecules. 11 (5), 726(2021).
- Gómez-Varela, D., Xian, F., Grundtner, S., Sondermann, J. R., Carta, G., Schmidt, M. Increasing taxonomic and functional characterization of host-microbiome interactions by DIA-PASEF metaproteomics. Front Microbiol. 14, 1258703(2023).
- Jagtap, P. D., et al. BAL fluid metaproteome in acute respiratory failure. Am J Respir Cell Mol Biol. 59 (5), 648-652 (2018).
- Masson, L., Wilson, J., Amir Hamzah, A. S., Tachedjian, G., Payne, M. Advances in mass spectrometry technologies to characterize cervicovaginal microbiome functions that impact spontaneous preterm birth. Am J Reprod Immunol Microbiol. 90 (2), e13750(2023).
- Bankvall, M., et al. Metataxonomic and metaproteomic profiling of the oral microbiome in oral lichen planus - a pilot study. J Oral Microbiol. 15 (1), 2161726(2023).
- Kruk, M. E., et al. An integrated metaproteomics workflow for studying host-microbe dynamics in bronchoalveolar lavage samples applied to cystic fibrosis disease. mSystems. 9 (7), e0092923(2024).
- Do, K., et al. A novel clinical metaproteomics workflow enables bioinformatic analysis of host-microbe dynamics in disease. mSphere. 9 (6), e00793-e00823 (2024).
- Batut, B., et al. Community-driven data analysis training for biology. Cell Syst. 6 (6), 752-758.e1 (2018).
- Hiltemann, S., et al. Galaxy Training: A powerful framework for teaching. PLoS Comput Biol. 19 (1), e1010752(2023).
- Galaxy Community. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 52 (W1), W83-W94 (2024).
- Blankenberg, D., et al. Dissemination of scientific software with Galaxy ToolShed. Genome Biol. 15 (2), 403(2014).
- Blank, C., et al. Disseminating metaproteomic informatics capabilities and knowledge using the Galaxy-P framework. Proteomes. 6 (1), E7(2018).
- Mehta, S., et al. A Galaxy of informatics resources for MS-based proteomics. Expert Rev Proteomics. 20 (11), 251-266 (2023).
- Armengaud, J. Metaproteomics to understand how microbiota function: The crystal ball predicts a promising future. Environ Microbiol. 25 (1), 115-125 (2023).
- Boylan, K. L., et al. A feasibility study to identify proteins in the residual Pap test fluid of women with normal cytology by mass spectrometry-based proteomics. Clin Proteomics. 11 (1), 30(2014).
- Afiuni-Zadeh, S., et al. Evaluating the potential of residual Pap test fluid as a resource for the metaproteomic analysis of the cervical-vaginal microbiome. Sci Rep. 8 (1), 10868(2018).
- Rauniyar, N., Yates, J. R. Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res. 13 (12), 5293-5309 (2014).
- Sivanich, M. K., Gu, T. -J., Tabang, D. N., Li, L. Recent advances in isobaric labeling and applications in quantitative proteomics. Proteomics. 22 (19-20), e2100256(2022).
- Potgieter, M. G., et al. MetaNovo: An open-source pipeline for probabilistic peptide discovery in complex metaproteomic datasets. PLoS Comput Biol. 19 (6), e1011163(2023).
- Vaudel, M., Barsnes, H., Berven, F. S., Sickmann, A., Martens, L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics. 11 (5), 996-999 (2011).
- Kim, S., Pevzner, P. A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 5, 5277(2014).
- Barsnes, H., Vaudel, M. SearchGUI: A highly adaptable common interface for proteomics search and de novo engines. J Proteome Res. 17 (7), 2552-2555 (2018).
- Vaudel, M., et al. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nature Biotechnol. 33 (1), 22-24 (2015).
- Tyanova, S., Temu, T., Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 11 (12), 2301-2319 (2016).
- Johnson, J. E., et al. Improve your Galaxy text life: The Query Tabular Tool. F1000Res. 7, 1604(2018).
- Wen, B., Wang, X., Zhang, B. PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res. 29 (3), 485-493 (2019).
- Wen, B., Zhang, B. PepQuery2 democratizes public MS proteomics data for rapid peptide searching. Nat Commun. 14 (1), 2213(2023).
- Pinter, N., et al. MaxQuant and MSstats in Galaxy enable reproducible cloud-based analysis of quantitative proteomics experiments for everyone. J Proteome Res. 21 (6), 1558-1565 (2022).
- Mesuere, B., Willems, T., Van Der Jeugt, F., Devreese, B., Vandamme, P., Dawyndt, P. Unipept web services for metaproteomics analysis. Bioinformatics. 32 (11), 1746-1748 (2016).
- Gurdeep Singh, R., et al. Unipept 4.0: Functional analysis of metaproteome data. J Proteome Res. 18 (2), 606-615 (2019).
- Verschaffelt, P., Collier, J., Botzki, A., Martens, L., Dawyndt, P., Mesuere, B. Unipept Visualizations: an interactive visualization library for biological data. Bioinformatics. 38 (2), 562-563 (2022).
- Huang, T., et al. MSstatsTMT: Statistical detection of differentially abundant proteins in experiments with isobaric labeling and multiple mixtures. Mol Cell Proteomics. 19 (10), 1706-1723 (2020).
- Choi, M., et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 30 (17), 2524-2526 (2014).
- Jagtap, P., et al. Workflow for analysis of high mass accuracy salivary data set using MaxQuant and ProteinPilot search algorithm. Proteomics. 12 (11), 1726-1730 (2012).
- Eng, J. K., Searle, B. C., Clauser, K. R., Tabb, D. L. A face in the crowd: recognizing peptides through database search. Mol Cell Proteomics. 10 (11), R111.009522(2011).
- Bihani, S., et al. Metaproteomics for coinfections in the upper respiratory tract: The case of COVID-19. Methods Mol Biol. 2820, 165-185 (2024).
- Jagtap, P., et al. A two-step database search method improves sensitivity in peptide sequence matches for metaproteomics and proteogenomics studies. Proteomics. 13 (8), 1352-1357 (2013).
- O'Bryon, I., Jenson, S. C., Merkley, E. D. Flying blind, or just flying under the radar? The underappreciated power of de novo methods of mass spectrometric peptide identification. Protein Sci. 29 (9), 1864-1878 (2020).
- Elias, J. E., Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 4 (3), 207-214 (2007).
- Kumar, D., Yadav, A. K., Dash, D. Choosing an optimal database for protein identification from tandem mass spectrometry data. Proteome Bioinformatics. 1549, 17-29 (2017).
- He, T., et al. Comparative evaluation of Proteome Discoverer and FragPipe for the TMT-based proteome quantification. J Proteome Res. 21 (12), 3007-3015 (2022).
- Searle, B. C., et al. Generating high quality libraries for DIA MS with empirically corrected peptide predictions. Nat Commun. 11 (1), 1548(2020).
- Easterly, C. W., et al. metaQuantome: An integrated, quantitative metaproteomics approach reveals connections between taxonomy and protein function in complex microbiomes. Mol Cell Proteomics. 18 (8 suppl 1), S82-S91 (2019).
- Lewis, M., et al. A Quantitative synthesis of early language acquisition using meta-analysis. , (2016).
- Bergmann, C., et al. Promoting replicability in developmental research through meta-analyses: Insights from language acquisition research. Child Dev. 89 (6), 1996-2009 (2018).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved