A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Gastric Mucosa Quantitative Polymerase Chain Reaction Analysis for Detecting Helicobacter pylori and Antibiotic Resistance
In This Article
Summary
A rapid and accurate method for H. pylori detection and drug resistance testing is very significant for efficiently eradicating H. pylori in clinical practice. This protocol aims to present a specific methodology involving gastric mucosa quantitative polymerase chain reaction (qPCR) for the rapid detection of H. pylori and antibiotic resistance.
Abstract
Helicobacter pylori is a main pathogen that infects nearly half of the global population and is threatening public health due to its increasing antibiotic resistance. Besides, Helicobacter pylori is also responsible for chronic gastritis, gastric and duodenal ulcers, gastric carcinoma, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma. Therefore, it is essential to perform a timely and accurate diagnosis of H. pylori and the determination of its antibiotic resistance. Nowadays, existing methods of H. pylori diagnosis mainly include the rapid urease test (RUT), the urea breath test (UBT), the serum antibody test, the antigen test, gastroscopy, and bacterial culture. However, bacteria could not be cultured through the first five detection methods, not to mention the detection of drug resistance. The bacterial culture is time-consuming, and antibiotic sensitivity tests cannot be carried out rapidly and routinely. In clinical settings, the swift and precise identification of H. pylori and its susceptibility to antibiotics is crucial for its effective elimination. The objective of this protocol is to outline a targeted approach utilizing quantitative polymerase chain reaction (qPCR) on gastric mucosal samples to expedite the diagnosis of H. pylori and assess its resistance to antimicrobial agents. qPCR was exploited to detect the ureA gene for H. pylori infection and mutations in the 23S rRNA and gyrA genes associated with resistance to clarithromycin and quinolones, respectively. Currently, there remain challenges in gastric mucosa qPCR due to the lack of standard operating procedures. Therefore, it is essential to share methodologies with experimental details to ensure accurate communication of experimental procedures, contributing to gold-standard protocols that enable greater transparency. Overall, this protocol offers an economical and expeditious alternative to conventional methods for assessing H.pylori infection and its resistance to antibiotics through the application of quantitative polymerase chain reaction (qPCR) technology.
Introduction
H. pylori is a gram-negative bacteria that can survive in the presence of a low oxygen level. The organism belongs to several distinct genetic populations and shows high genetic diversity. Although the organism is usually spirally shaped, it could be changed to a rod1. It is a main pathogen that infects nearly 50% of the global population2. Mostly, infection occurs when patients remain healthy carriers during childhood, and symptoms manifest later in adulthood. It has been reported to be relative to chronic gastritis, gastric and duodenal ulcers, gastric carcinoma, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma3. Differences in the manifestations of H. pylori infection could stem from the pathogenicity elements of distinct bacterial strains, as well as the attributes of the host and their dietary patterns4. The research indicates that men who engage in smoking and alcohol consumption are at a higher risk of contracting H. pylori infections5. The efficacy of eliminating H. pylori for preventing stomach cancer and precancerous lesions has been verified in several studies6,7. Therefore, H. pylori eradication was advised as a preventative measure by the World Health Organization (WHO) International Agency for Research on Cancer8.
The rapid and accurate diagnosis of H. pylori infection is a critical part of treatment for most individuals suffering from asymptomatic dyspepsia. The diagnosis of H. pylori involves a combination of both invasive and non-invasive approaches. The invasive techniques typically encompass endoscopy for direct visualization, histological analysis of tissue samples, the rapid urease test (RUT) to detect bacterial activity, and culturing of the bacteria. On the other hand, non-invasive strategies comprise the urea breath test (UBT), which measures bacterial metabolism; the stool antigen test (SAT), which detects bacterial proteins in feces; serological tests that look for antibodies in the blood; and molecular diagnostics that use genetic material for detection. While each of these diagnostic methods comes with its own set of benefits and drawbacks, in the realm of clinical practice, there is no single method that is universally acknowledged as the ultimate benchmark9.
Currently, the primary treatment for H. pylori infections is antibiotics. However, the success rates of antimicrobial therapies aimed at eradicating H. pylori have been on a decline, attributable to various factors. The predominant causes of treatment ineffectiveness are suspected to be the bacteria's resistance to antimicrobial agents and the adherence of patients to the prescribed regimen10. Specifically, strains of H. pylori that are resistant to clarithromycin have been the subject of extensive research11. The incidence of such resistance is on the rise in numerous nations. H. pylori resistance is mainly due to mutations in the variable region gene of 23S rRNA, which causes conformational changes in the ribosome. Consequently, the binding site for clarithromycin changes, leading to a weakened affinity between H. pylori and clarithromycin, preventing the inhibition of bacterial protein synthesis12. Mutations at positions A2143G, A2142G, and A2142C within the 2.9 kb segment of the 23S rRNA gene are known to confer resistance to clarithromycin. The spread of resistance to fluoroquinolones has also been a focus of numerous investigations. Studies have documented resistance rates to levofloxacin at 34.5% in China and 22.1% in Italy13. Quinolone drugs mainly act on H. pylori's topoisomerase II by inhibiting enzyme activity, affecting DNA synthesis and replication and secondary structure, thereby achieving antibacterial purposes. If mutations occur in the genes encoding the topoisomerase subunits, gyrA, and gyrB, it will prevent the binding of antibiotics such as levofloxacin and the enzyme, resulting in the inability to inhibit the replication of the H. pylori genome, thus causing resistance. Among these, the hotspots of mutations in the gyrA gene are concentrated at amino acids 87 and 91, while mutations in the gyrB gene occur less frequently and are often accompanied by mutations in gyrA14. The mutation loci of the levofloxacin resistance gene mainly include the six mutation sites (A260T, C261A, T261G, G271A, G271T, A272G) located in the gyrA gene. The identification of resistance mechanisms stemming from genetic alterations has prompted a progressive transition in the detection of H. pylori, moving away from culture-based methods towards molecular diagnostic techniques.
UBT and SAT are the most commonly chosen non-invasive tests, but they cannot provide drug susceptibility information. Consequently, the development of a swift and comprehensive technique for detecting H. pylori and assessing its resistance to medications is crucial for its effective elimination in clinical settings15. Among the molecular detection techniques, quantitative polymerase chain reaction (qPCR) has seen significant advancements. Unlike standard PCR, qPCR eliminates the need for gel electrophoresis and enables the precise quantification of DNA or RNA by incorporating specific primers and probes during the annealing phase. Commercially available qPCR kits now offer the capability to identify H. pylori infections and resistance to drugs16.
Basically, there is an immediate clinical need for a diagnostic approach that is both potent and comprehensive, capable of detecting H. pylori infections and assessing drug resistance concurrently. We adopted gastric mucosa qPCR analysis for H. pylori detection and antibiotic resistance using different primer probes.
Access restricted. Please log in or start a trial to view this content.
Protocol
The existing study was conducted in conformity with ethical considerations established by the ethical committee of Guangdong Provincial People's Hospital, Southern Medical University, Guangzhou, China (Approval Number: KY2024-1115-01). Patients aged from 18 to 60 were enrolled in this study. For this study, participants were excluded if they had recently taken antibiotics, antibacterial herbal remedies, proton pump inhibitors (PPIs), or H2 receptor antagonists within the 2 weeks prior to testing. Additionally, individuals who had undergone anti-H. pylori therapy within the last 3 months, or those with significant cardiac, hepatic, or renal issues, severe neuropathy, or psychiatric disorders were not eligible for participation. Those with contraindications for gastroscopy examination, such as gastrointestinal perforation, advanced age, unstable vital signs, etc., were excluded. The study also did not include pregnant or breastfeeding women. The informed consent form of the subjects mainly includes the following contents: (1) research background and purpose; (2) Who is unsuitable to participate in this research project? (3) What are the requirements for participating in the research? (4) Possible benefits of participating in research; (5) Possible adverse reactions, risks, and discomfort; (6) Privacy protection; (7) Subject rights; (8) Subject declaration; (9) Researcher statement, etc.
1. Sampling of the gastric mucosa
- Preparation of sampling consumables
- Prepare the sampling tube containing the storage solution.
- Prepare gastroscope biopsy forceps.
- Preparation of sampling objects
- Collect specimens from patients with chronic gastritis, gastric ulcer, duodenal ulcer, gastric cancer, or suspected patients. Collect samples maintaining the aseptic procedures.
NOTE: The collected specimens included gastric mucosal tissue from patients or suspected patients. - Confirm the drug history.
- Ensure that no antibiotics and bismuth for H. pylori are taken by the patient within 4 weeks and that no acid inhibitor is taken within 2 weeks.
- Request patients taking anticoagulant drugs (warfarin, aspirin, clopidogrel, etc.) to stop taking medication for 5-7 days.
- Collect specimens from patients with chronic gastritis, gastric ulcer, duodenal ulcer, gastric cancer, or suspected patients. Collect samples maintaining the aseptic procedures.
- Collection of specimens
- Sampling site
- According to the requirements of the new Sydney system, take biopsy samples from the gastric antrum, the large and small bend of the gastric body, and the gastric corner (a total of 5 points). The specific sites are shown in Figure 1. A small bend of the pyloric sinus 2-3cm from the pyloric ring (A1) and a big bend (A2); 4 cm from the stomach (B1) and a large bend 8 cm from the cardia (B2); gastric corner (IA).
- According to Expert Consensus on the Standardization of Digestive Endoscopic Biopsy and Pathological Examination in China (Draft), take biopsy samples from the small side of the gastric sinus 5 cm from the pylorus (adjacent to the gastric corner) or the large side of the gastric corner (biopsy 1-2 pieces).
- Description of the collection location of special samples
- For patients under any treatment, after a withdrawal for 4 weeks, collect gastric antrum and gastric body samples simultaneously.
- For patients with gastric cancer, collect tissue from the large curvature of the stomach but not the tumor tissue.
- For patients with gastric ulcers, collect the gastric antrum and gastric ulcer edge tissue. Do not collect the bottom of the ulcer.
- For patients with atrophic gastritis, collect gastric antrum and gastric body. Do not collect samples from gastric mucosal atrophy and intestinal metaplasia areas.
- Sampling steps
- Open the biopsy clamp valve and move it slowly to the dominant hand. When the head of the forceps is in the field of vision, open the clamp flap and manipulate the endoscope at the gastric mucosa of the sampling site.
- Apply a slight pressure, close the biopsy forceps, and clamp the gastric mucosa tissue (diameter: 0.5-1 mm, about 5-10 mg/block). Extract the tissue samples with the pliers in the sampling tube (including preservation fluid) to complete the site.
- Continue to complete the sampling of other sites and place them in the same sampling tube (including preservation solution).
- Complete the sampling of all sites, tighten the sampling tube cover, and seal to prevent drying. Label the tubes with a unique identification number on the outside.
- Submit the collected samples for testing as soon as possible. Do not place the samples at room temperature (RT) for more than 24 h. Avoid repeated freezing and thawing of clinical specimens during long-distance transportation. If a temperature of -20 ± 5 °C cannot be ensured, transport them at 0-8 °C. Store the samples for a period of five months at -20 ± 5 °C, and for long-term storage, keep them below -70 °C.
- Precautions
- When sampling, press the biopsy forceps as deep as possible to reach the whole layer of mucosa.
- When removing the biopsy forceps and opening the biopsy, be gentle to prevent mucosal tears.
- Sampling site
2. Nucleic acid extraction
- Adjust the temperature of the metal bath to 100 °C in advance; if the sample is frozen, remove it and restore it to RT.
- Mix the lysate well to suspend the iminodiacetic acid resin. Take 100 µL of lysate into the centrifugal tube with the filter element, or directly add the gastric mucosa tissue into the centrifuge tube with 200 µL of lysate and mix with vortex oscillation.
- Place the centrifuge tube in a metal bath at 100 °C for 10 min, then remove and cool to RT.
- Centrifuge at 9500 g for 5-10 min. Carefully draw the supernatant into another sterilized centrifuge tube and mark it.
- Immediately conduct the following experiment or temporarily store it at 4 °C for inspection.
3. qPCR detection of H. pylori and drug-resistance genes (clarithromycin and quinolones)
- Remove the primer probe, the mixed enzyme solution, and the detection buffer from the kit. Melt all the components on ice or 2-8 °C, and mix them with slight shaking and instantaneous centrifugation at low speed.
- Mix 12.5 µL of detection buffer + 7.0 µL of primer probe + 0.5 µL of enzyme solution. Calculate the amount of each reagent first, add to the appropriate volume centrifuge tube, mix well, and centrifuge briefly.
NOTE: The total amount of the PCR reaction solution should be calculated by including the number of test samples, one strong positive control, one weak positive control, one negative control, and one PCR negative control. - After the PCR reaction solution is configured, add 20.0 µL of the PCR reaction solution to each PCR reaction well. Then, add 5.0 µL of nucleic acid to the PCR reaction well with the PCR reaction solution. Add no sample or nucleic acid to the PCR negative control well.
- Put the reaction tube into the fluorescent PCR thermocycler and set the cycle parameters as follows: first, heat the reaction mixture at 42 °C and 95 °C for 2 min; second, denature for 10 s at 95 °C, anneal and extend for 45 s at 58 °C, repeat for 40 cycles.
- Collect the fluorescence signals as FAM (23S rRNA gene mutation), HEX/VIC (gyrA gene mutation), ROX (H. pylori), and CY5 (internal standard), and data at 58 °C.
- After the reaction, save the data and analyze it using specific qPCR software.
Access restricted. Please log in or start a trial to view this content.
Results
Assessment of H. pylori Infection and Antibiotic Resistance in Gastric Tissue via qPCR
The qPCR assays for identifying H. pylori were conducted by targeting the ureA gene, and antibiotic resistance was ascertained by examining specific mutations in the 23S rRNA and gyrA genes (Table 1). The quality assurance data, represented by the CT values across all three groups, fell within acceptable limits, signifying that...
Access restricted. Please log in or start a trial to view this content.
Discussion
Traditional testing such as RUT, UBT, histology, culturing as well as serology are exploited for the detection of H. pylori. Each diagnostic approach offers distinct advantages and faces specific challenges depending on the clinical context17. The cultivation of H. pylori from gastric mucosal biopsies is often considered the benchmark for diagnosis. Nonetheless, this method is labor-intensive, and its accuracy is constrained by technical challenges, the conditions required for in...
Access restricted. Please log in or start a trial to view this content.
Disclosures
None.
Acknowledgements
This work was supported by grants from the NSFC Incubation Program of GDPH (8220080645) and the Guangdong Provincial Medical Science and Technology Research Fund Project (A2024108).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Bath Incubator | ALLSHENG | MK2000-2 | Provide a constant temperature environment |
| Biosafety cabinet | Haier | HR1500-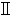 B2 B2 | |
| Centrifuge | Thermo Fisher Scientific | THERMO ST16R | Centrifuge the residual liquid off the wall of the tube. |
| Chelex-100 | sigma | C7901 | Resin |
| Gastroscope biopsy forceps | Boston Scientific Corporation | Sampling of the gastric mucosa | |
| Helicobacter pylori 23S rRNA gene and gyrA gene mutation detection kit | Jiangsu Mole Bioscience | CFDA 20223400137 | qPCR detection kit for H. pylori and drug-resistance genes (clarithromycin and quinolones) |
| Nucleic acid extraction reagent | Jiangsu Mole Bioscience | SEDA 20150076 | For DNA extraction |
| SDS Software | Applied Biosystems | 7300/7500 | Data analysis |
| Thermocylcer | Thermo Fisher Scientific | ABI 7500 | For qPCR detection of H. pylori and drug-resistance genes (clarithromycin and quinolones) |
| Vortex mixer | JOANLAB | VM-5005 | For mixing reagent |
References
- Sharndama, H. C., Mba, I. E. Helicobacter pylori: an up-to-date overview on the virulence and pathogenesis mechanisms. Braz J Microbiol. 53 (1), 33-50 (2022).
- Ravikumara, M. Helicobacter pylori in children: think before you kill the bug. Therap Adv Gastroenterol. 16 (6), 1-10 (2023).
- Reshetnyak, V. I., Burmistrov, A. I., Maev, I. V. Helicobacter pylori: Commensal, symbiont or pathogen. World J Gastroenterol. 27 (7), 545-560 (2021).
- Al-Ouqaili, M. T. S., et al. Study of vacuolating cytotoxin A (vacA) genotypes of ulcerogenic and non-ulcerogenic strains of Helicobacter pylori and its association with gastric disease. Saudi J Biol Sci. 30 (12), 103867(2023).
- Hussein, R. A., Al-Ouqaili, M. T. S., Majeed, Y. H. Association between alcohol consumption, cigarette smoking, and Helicobacter pylori infection in Iraqi patients submitted to gastrointestinal endoscopy. J Emerg Med Trauma Acute Care. 6, 12(2022).
- Thrift, A. P., Wenker, T. N., El-Serag, H. B. Global burden of gastric cancer: Epidemiological trends, risk factors, screening and prevention. Nat Rev Clin Oncol. 20 (5), 338-349 (2023).
- Liou, J. M., et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: The Taipei global consensus. Gut. 69 (12), 2093-2112 (2020).
- Gao, T. Y., et al. Cancer burden and risk in the Chinese population aged 55 years and above: A systematic analysis and comparison with the USA and Western Europe. J Glob Health. 14, 04014(2024).
- Hussein, R. A., Al-Ouqaili, M. T. S., Majeed, Y. H. Detection of Helicobacter pylori infection by invasive and non-invasive techniques in patients with gastrointestinal diseases from Iraq: A validation study. PLoS One. 16 (8), e0256393(2021).
- Thung, I., et al. Review article: The global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 43 (4), 514-533 (2016).
- Zhang, Y., et al. Mutations in the antibiotic target genes related to clarithromycin, metronidazole and levofloxacin resistance in Helicobacter pylori strains from children in China. Infect Drug Resist. 13 (1), 311-322 (2020).
- Saranathan, R., et al. Helicobacter pylori Infections in the Bronx, New York: Surveying antibiotic susceptibility and strain lineage by whole-genome sequencing. J Clin Microbiol. 58 (3), e01591-e01619 (2020).
- Malfertheiner, P., et al. Management of Helicobacter pylori infection - The Maastricht IV/ Florence consensus report. Gut. 61 (5), 646-664 (2012).
- Palma, G. Z. D., et al. Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from Buenos Aires city. Microb Drug Resist. 23 (3), 351-358 (2017).
- Zhang, L., Zhao, M., Fu, X. S. Gastric microbiota dysbiosis and Helicobacter pylori infection. Front Microbiol. 14, 1153269(2023).
- Peng, X., et al. Gastric juice-based real-time PCR for tailored Helicobacter Pylori treatment: A practical approach. Int J Med Sci. 14 (6), 595-601 (2017).
- Weingart, V., et al. Sensitivity of a novel stool antigen test for detection of Helicobacter pylori in adult outpatients before and after eradication therapy. J Clin Microbiol. 42 (3), 1319-1321 (2004).
- Chi, W. J., et al. A rapid and high-throughput multiplex genetic detection assay for detection, semi-quantification and virulence genotyping of Helicobacter pylori in non-invasive oral samples. Front Cell Infect Microbiol. 13, 1267288(2023).
- Zhang, Y. M., et al. Validation of a high-throughput multiplex genetic detection system for Helicobacter pylori identification, quantification, virulence, and resistance analysis. Front Microbiol. 7, 1401(2016).
- Gisbert, J. P. Empirical or susceptibility-guided treatment for Helicobacter pylori infection? A comprehensive review. Therap Adv Gastroenterol. 13, 1756284820968736(2020).
- Bénéjat, L., et al. Real-time PCR for Helicobacter pylori diagnosis. The best tools available. Helicobacter. 23 (5), e12512(2018).
- Vasapolli, R., et al. Real-time assessment of H. pylori infection to guide molecular antibiotic resistance testing: A combined endoscopy-gastric juice analysis approach. Aliment Pharmacol Ther. 61 (3), 465-471 (2025).
- Hussein, R. A., Al-Ouqaili, M. T. S., Majeed, Y. H. Detection of clarithromycin resistance and 23SrRNA point mutations in clinical isolates of Helicobacter pylori isolates: Phenotypic and molecular methods. Saudi J Biol Sci. 29 (1), 513-520 (2022).
- Wang, Y. H., et al. A systematic review and meta-analysis of genotypic methods for detecting antibiotic resistance in Helicobacter pylori. Helicobacter. 23 (2), e12467(2018).
- Egli, K., et al. Comparison of the diagnostic performance of qPCR, Sanger sequencing, and whole-genome sequencing in determining clarithromycin and levofloxacin resistance in Helicobacter pylori. Front Cell Infect Microbiol. 10, 596371(2020).
- Liu, Y., et al. Relationship between Helicobacter pylori infection and colorectal polyp/colorectal cancer. World J Gastrointest Surg. 16 (4), 1008-1016 (2024).
- Oliver, A. S., Wu, F., Chen, Y. The role of gastric microbiota in gastric cancer. Gut Microbes. 11 (5), 1220-1230 (2020).
- Hulten, K. G., et al. Comparison of culture with antibiogram to next-generation sequencing using bacterial isolates and formalin-fixed, paraffin embedded gastric biopsies. Gastroenterology. 161 (5), 1433-1442 (2021).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved