A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Determination of Zeta Potential via Nanoparticle Translocation Velocities through a Tunable Nanopore: Using DNA-modified Particles as an Example
In This Article
Summary
Here we use a polyurethane tunable nanopore integrated into a resistive pulse sensing technique to characterize nanoparticles surface chemistry via the measurement of particle translocation velocities, which can be used to determine the zeta potential of individual nanoparticles.
Abstract
Nanopore technologies, known collectively as Resistive Pulse Sensors (RPS), are being used to detect, quantify and characterize proteins, molecules and nanoparticles. Tunable resistive pulse sensing (TRPS) is a relatively recent adaptation to RPS that incorporates a tunable pore that can be altered in real time. Here, we use TRPS to monitor the translocation times of DNA-modified nanoparticles as they traverse the tunable pore membrane as a function of DNA concentration and structure (i.e., single-stranded to double-stranded DNA).
TRPS is based on two Ag/AgCl electrodes, separated by an elastomeric pore membrane that establishes a stable ionic current upon an applied electric field. Unlike various optical-based particle characterization technologies, TRPS can characterize individual particles amongst a sample population, allowing for multimodal samples to be analyzed with ease. Here, we demonstrate zeta potential measurements via particle translocation velocities of known standards and apply these to sample analyte translocation times, thus resulting in measuring the zeta potential of those analytes.
As well as acquiring mean zeta potential values, the samples are all measured using a particle-by-particle perspective exhibiting more information on a given sample through sample population distributions, for example. Of such, this method demonstrates potential within sensing applications for both medical and environmental fields.
Introduction
Functionalized nanoparticles are becoming increasingly popular as biosensors in both medical and environmental fields. The ability to alter a nanoparticle's surface chemistry, with DNA, for example, is proving useful for targeted drug delivery systems1 and monitoring DNA-protein interactions2-4. An increasingly common nanoparticle property being utilized in bioassays and in the delivery of therapeutics is superparamagnetism5. Superparamagnetic particles (SPPs) are extremely useful in identifying and removing specific analytes from complex mixtures and can do so with the simple use of a single magnet. Once removed, the analyte-bound particles can be characterized and analyzed fit for purpose.
Previous methods used for the detection and characterization of nanoparticles include optical techniques such as dynamic light scattering (DLS), otherwise known as photon correlation spectroscopy. Although a high throughput technique, DLS is limited to being an averaging based technique and when analyzing multimodal samples without the addition of specialist software, the larger particles will produce a much more dominant signal, leaving some of the smaller particles completely unnoticed6,7. Particle-by-particle characterization techniques are therefore much more favorable to analyze nanoparticle and functionalized nanoparticle systems.
RPS based technologies are based around applying an electric field to a sample and monitoring the transportation mechanism of the particles through a synthetic or biological nanopore. A relatively recent nanoparticle detection and characterization technique based on RPS is tunable resistive pulse sensing (TRPS)8-16. TRPS is a two-electrode system separated by an elastomeric, tunable pore membrane. A tunable pore method allows for analytes of a range of shape17 and size to be measured via their transport mechanisms through the pore. Tunable pores have previously been used for the detection of small particles (70-95 nm diameter) producing comparable results to other techniques such as transmission electron spectroscopy (TEM)10. When an electric field is applied, an ionic current is observed and as particles/molecules pass through the pore, they temporarily block the pore, causing a reduction in the current that can be defined as a 'blockade event'. Each blockade event is representative of a single particle so that each particle within a sample can be characterized individually based on the blockade magnitude, Δ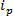 , and full width half-maximum, FWHM, as well as other blockade properties. Analyzing individual particles as they pass through a nanopore is advantageous for multimodal samples as TRPS can successfully and effectively distinguish a range of particle sizes amongst a single sample. Tunable resistive pulse sensing completes size10, zeta potential12,18 and concentration15 measurements simultaneously in a single run and can therefore still differentiate samples of similar, if not the same size by their surface charge19; an advantage over alternative sizing techniques.
, and full width half-maximum, FWHM, as well as other blockade properties. Analyzing individual particles as they pass through a nanopore is advantageous for multimodal samples as TRPS can successfully and effectively distinguish a range of particle sizes amongst a single sample. Tunable resistive pulse sensing completes size10, zeta potential12,18 and concentration15 measurements simultaneously in a single run and can therefore still differentiate samples of similar, if not the same size by their surface charge19; an advantage over alternative sizing techniques.
Zeta potential is defined as the electrostatic potential at the plane of shear20, and is calculated from particle velocities as they traverse a pore19. Zeta potential measurements of individual particles thus gives insight into the translocation mechanisms and behavior of nanoparticle systems in solution, valuable information for the future of nanoparticle assay designs for a range of applications. Particle-by-particle analysis of such nature also allows for the spread and distribution of zeta potential values amongst a sample population to be explored, allowing for more information on reaction kinetics (single-stranded to double-stranded DNA, for example) and particle stabilities in solution to be attained.
Here, we describe a technique that detects and characterizes both unmodified and DNA-modified SPP surfaces. The protocol described herein is applicable to a range of inorganic and biological nanoparticles, but we demonstrate the procedure using DNA-modified surfaces due to their wide range of applications. The technique allows the user to distinguish between single-stranded and double-stranded DNA targets on a nanoparticle surface, based on particle translocation velocities through a pore system and thus their zeta potentials.
Protocol
1. Making the Phosphate Buffered Saline with Tween-20 (PBST) Buffer
- Dissolve one PBS tablet (0.01 M phosphate buffer, 0.0027 M Potassium Chloride, 0.137 M Sodium Chloride, pH 7.4) in 200 ml deionized water (18.2 MΩ cm).
- Add 100 µl (0.05 (v/v)%) Tween-20 to the 200 ml buffer solution as a surfactant.
2. Preparing the Carboxyl Polystyrene Particle Standards
- Vortex the calibration particles for 30 sec before sonication for 2 min at 80 watts to create monodispersity of the particles.
- Dilute the calibration particles 1 in 100 to a concentration of 1x1010 particles/ml in PBST buffer and vortex for 30 sec.
3. Preparing Streptavidin Coated Particles
- Vortex the particles for 30 sec before sonication for 2 min at 80 watts to ensure monodispersity.
- Dilute the streptavidin coated particles 1 in 100 in PBST buffer to achieve a resulting concentration of 1x109 particles/ml and vortex for 30 sec.
Note: A typical sample volume is 200 µl. For example, if investigating five DNA concentrations prepare 1 ml of diluted streptavidin coated particles.
4. Preparation of Oligonucleotides
- Reconstitute oligonucleotides with deionized water to a resulting concentration of 100 µM.
5. Addition of Capture Probe (CP) DNA to the Streptavidin Coated Particles
- Prior to DNA binding, vortex the streptavidin coated particles (200 µl sample volume) for 30 sec followed by a 2 min sonication at 80 watts.
- Based on the binding capacity provided by the supplier (4,352 pmol/mg), add the appropriate concentration of DNA to the particles for resulting concentrations of 10, 20, 30, 40, 47, 95, 140, and 210 nM DNA.
- Vortex the samples for 10 sec and place on a rotary wheel at room temperature for 30 min to allow for the DNA to bind to the particle surfaces via a streptavidin-biotin interaction.
- Once the capture DNA has been added and incubated with the streptavidin coated particles, remove the excess DNA in solution via magnetic separation by placing the samples onto a magnetic rack for 30 min.
- Remove the supernatant, taking care not to disturb the newly formed cluster of particles closest to the magnet, and replace with the same volume of new PBST buffer.
6. Hybridizing Complementary DNA to the CP-particles
- Add the required amount of target DNA (in excess at 500 nM) to ensure the maximum possible target binding was reached.
- Vortex the samples for 10 sec and place on a rotary wheel at room temperature for 30 min.
- Once the hybridization is complete, remove the excess target DNA via magnetic separation by placing the samples onto a magnetic rack for 30 min.
- Remove the supernatant, taking care not to disturb the newly formed cluster of particles closest to the magnet, and replace with the same volume of new PBST buffer.
- Repeat steps 6.1 to 6.4 for duplicate samples and place these samples on a rotary wheel at room temperature for 16 hours to investigate DNA hybridization times.
7. TRPS Setup
- Plug in the instrument into a computer system with software in place.
- Calibrate the initial stretch using a caliper.
- Measure the distance between the outside of two parallel jaws.
- Input into the software by typing the stretch in the 'stretch' field in the 'Instrument Settings' tab and clicking 'Calibrate stretch' underneath the tab.
- Laterally fit a polyurethane nanopore membrane of appropriate sizing for analysis onto the jaws with the nanopore ID number facing upward. Then, stretch the jaws to the stretch required for analysis using the stretch adjustment handle on the side of the instrument. Stretch the jaws between 43 and 48 mm.
Note: The exact value of the stretch is determined alongside applied voltage so that calibration particle blockades are at least 0.3 nA in size. The stretch is already inputted into the software in step 7.2 and will automatically adjust as the jaws are stretched. - Place 80 µl of PBST buffer in the lower fluid cell, beneath the nanopore, ensuring there are no bubbles present that may affect the measurement. If there are bubbles seen, remove and replace the buffer.
- Click the upper fluid cell into place and place 40 µl of buffer into it, again ensuring there are no bubbles present. If bubbles are present in the upper fluid cell, remove them by replacing the liquid.
- When a reproducible baseline current has been reached from replacing the upper fluid cell with buffer, add 40 µl of the sample to the upper fluid cell and measure by clicking 'start' in the 'Data Acquisition' tab on the software screen.
Note: The data acquisition is completed at a frequency of 50 kHz with a blockade magnitude lower limit of 0.05 nA, although this can be altered using the software via the 'Analyse Data' tab (under 'Analysis Settings' and 'Resistive Blockades'). - Place a Faraday cage over the top of the fluid cell system to reduce electrical background noise on the measurements.
- Use a variable pressure module (VPM) to apply a pressure or vacuum to the samples.
- To apply an external pressure connect the nozzle to the upper fluid cell, then rotate the pressure arm and click into place (depending on whether a positive pressure (PRE) or a vacuum (VAC) will be applied).
- Apply pressure in a 'cm' or 'mm' scale using the pressure stage knob situated on the top of the VPM. Press the knob down to apply pressure on the 'cm' scale and pull it upward to apply pressure on the 'mm' scale.
8. Preparing Samples for TRPS Analysis
- Vortex samples for 30 sec and sonicate for 2 min at 80 watts prior to TRPS analysis.
9. Calibrating the Nanopore for Zeta Analysis
- After placing 40 µl calibration particles (1x1010 particles/ml) into the upper fluid cell, complete a TRPS measurement (setup as in section 7) at 3 applied voltages. Alter the voltage by clicking on the '+' and '-' buttons on the voltage scale in the 'Instrument Settings' tab on the software.
- Check that the 3 voltages return background currents of approximately 140, 110, and 80 nA. Ensure that at the medium voltage the calibration particles produce an average blockade magnitude of at least 0.3 nA.
- Apply a pressure so the average full width half maximum (FWHM) durations of the calibration particles are at least 0.15 msec. Do this manually using the pressure arm attached to the variable pressure module. Select pressure (PRE) or vacuum (VAC) by rotating the arm until it clicks in the desired position and apply accordingly following set up instructions in step 7.8.2. Once these conditions have been achieved, start the run by clicking 'start' on the software in the 'Data Acquisition' tab.
- Complete the run by pressing 'stop' in the 'Data Acquisition' tab when at least 500 particles have been measured (see 'Particle Count' at the bottom of the software screen during the measurement) and the run has exceeded 30 sec (see 'Run Time' also toward the bottom of the screen).
- Calibrate the system by completing a calibration run as described every time a new nanopore is introduced or for each new day of analysis by completing step 9.1-9.4.
10. Running a Sample
- Run the samples at the highest or second highest voltage as the calibration samples at ensuring a similar (±10 nA), if not the same, baseline current.
- Once the appropriate baseline current is achieved, replace the electrolyte in the upper fluid cell with 40 µl of sample. When a sample is introduced, blockades will be seen on the signal trace. Start the sample run by clicking 'start' in the 'Data Acquisition' tab and record a minimum of 500 particles (check 'Particle Count' situated under the signal trace) and ensure the run time is a minimum of 30 sec (see 'Run Time' also situated below the signal trace).
- To complete the measurement, click 'stop' in the 'Data Acquisition tab' and save the data file.
- To save the file, input the file information in the following format; 'Investigation' is the folder the file will be saved in, 'Nanopore ID' is the serial number of the pore being used, 'Part #' is the type of pore (i.e., NP150/NP200), 'Sample ID' is the name of the sample, 'Calibration or sample' details whether it is a calibration or sample measurement, 'Dilution' is used if the sample was diluted (type 100 if the sample was diluted 100-fold), 'Pressure' is the applied pressure to the sample (in cm - see section 7.8), 'Electrolyte ID' is the name of the buffer the sample is made up in, and 'Notes' are any personal notes about the sample or run.
- Between each sample run, wash the system by placing 40 µl of PBST buffer into the upper fluid cell several times and applying various pressures (usually at -10, -5 cm (vacuum), and 5 and 10 cm (positive pressure)) until no more blockade events are present, ensuring there are no residual particles remaining in the system and therefore no cross contamination between samples. Run samples in triplicate with this wash step completed between each repeat sample run as well as between different samples.
Results
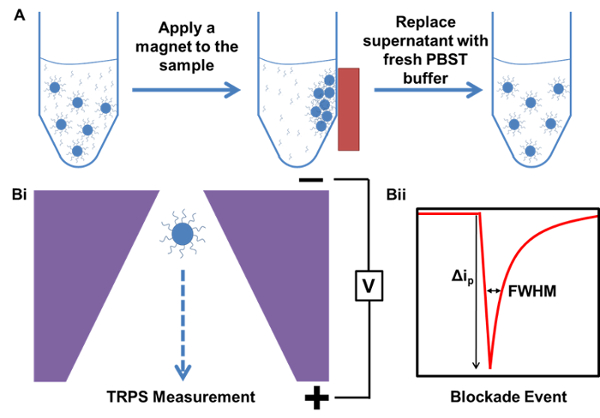
Figure 1. Schematic representation of the processes of magnetic purification and a TRPS measurement. A) Example of magnetic purification of sample starting with a sample containing excess, unbound capture probe DNA. B) TRPS measurement example i) Particle passing through the nanopore and ii) Blockade event produced from particle temporarily occluding ions in the pore...
Discussion
The calculation for the zeta potential used a calibration based method related to work by Arjmandi et al.21. The duration of the translocation of particles as they traverse a nanopore is measured as a function of applied voltage, using an average electric field and particle velocities over the entirety of a regular conical pore. The electrophoretic mobility is the derivative of 1/T (where T is the blockade duration) with respect to voltage, multiplied by the square of the sensing zone lengt...
Disclosures
E.L.C.J.B. is supported by Izon Science Ltd.
Acknowledgements
The authors thank Izon Science Ltd for their support. The work was supported by the European Commission for Research (PCIG11-GA-2012-321836 Nano4Bio).
Materials
| Name | Company | Catalog Number | Comments |
| Phosphate buffered Saline (PBS) | Sigma Aldrich, UK | P4417 | 1 tablet dissolved in 200 ml deionized water to make buffer solution. |
| Tween-20 | Sigma Aldrich, UK | P1379 | 0.05% (v/v) in PBS buffer as a surfactant |
| Carboxyl polystyrene nanoparticles | Bangs Laboratories, US | CPC200 | Nominal diamter of 220 nm, raw concentration of 1 x 1012 particles/ml, specific surface charge of 86 µeq/g (equivalent to a surface charge density of 3.2 x 1019 C/nm2. |
| Streptavidin coated nanoparticles | Ademtech, France | 3121 | Batch had binding capacity of 4,352 pmol/mg (188 nM theoretical DNA binding capacity) at a raw concentration of 1.1 x 1011 particles/ml. |
| Biotinylated oligonucleotides | Sigma Aldrich, UK | VC00001 | Supplier spec: Reverse Phase 1 purification (0.05 Scale); Biotin modification at 3' end; Lyophilized powders reconstituted to 100 µM using deionized water, and diluted as required. Sequences: CP 5'ATGGTTAAACCTCACTAC GCGTGGC[Btn]3' |
| Standard olignonucleotides | Sigma Aldrich, UK | VC00001 | Supplier spec: Reverse Phase 1 purification (0.05 Scale); Lyophilized powders reconstituted to 100 µM using deionized water, and diluted as required. Sequences of DNA targets: Fully complementary - 5'GCCACGCGTAGTGA GGTTTAACCAT3', Middle binding - 5'GTAGTGAGGT3', End binding - 5'GTTTAACCAT3', Partially complementary overhanging - 5'GTGAGGTTTAACCAT TTTTTTTTTTTTTTT3'. |
| Izon qNano | Izon Science, NZ | Inherent pressure on system of 47 Pa | |
| Izon Variable Pressure Module (VPM) | Izon Science, NZ | Each 'cm' of pressure is equivalent to approximately 100 Pa. | |
| Polyurethane nanopore membranes | Izon Science, NZ | NP150 | Analyte size range 60-480 nm, pore diameter of calculated to be 799 nm at a 45 mm stretch. |
| Magrack 6 | GE Healthcare, UK | 28-9489-64 | |
| Sonic Bath | Fisher Scientific, UK | 10692353 | 80 Watts |
| Vortexer | IKA, Germany | 0003365000 | |
| Rotary Wheel | Labnet International, US | H5500-230 V |
References
- Alexander, C. M., Maye, M. M., Dabrowiak, J. C. DNA-capped nanoparticles designed for doxorubicin drug delivery. Chem Commun. 47 (12), 3418-3420 (2011).
- Billinge, E. R., Platt, M. Aptamer based dispersion assay using tunable resistive pulse sensing (TRPS). Anal Methods. 7 (20), 8534-8538 (2015).
- Bulyk, M. L. Protein Binding Microarrays for the Characterization of Protein-DNA Interactions. Adv Biochem Eng Biotechnol. 104, 65-85 (2007).
- Platt, M., Rowe, W., Knowles, J., Day, P. J., Kell, D. B. Analysis of aptamer sequence activity relationships. Integr Biol. 1 (1), 116-122 (2009).
- Ruiz-Hernández, E., Baeza, A., Vallet-Regí, M. Smart Drug Delivery through DNA/Magnetic Nanoparticle Gates. ACS Nano. 5 (2), 1259-1266 (2011).
- Murdock, R. C., Braydich-stolle, L., Schrand, A. M., Schlager, J. J., Hussain, S. M. Characterization of Nanomaterial Dispersion in Solution Prior to In Vitro Exposure Using Dynamic Light Scattering Technique. Toxicol Sci. 101 (2), 239-253 (2008).
- Hupfield, S., Holsaeter, A. M., Skar, M., Frantzen, C. B., Brandl, M. Liposome size analysis by dynamic/static light scattering upon size exclusion-/field flow fractionation. J Nanosci Nanotechnol. 6 (7), 3025-3031 (2006).
- Roberts, G. S., et al. Tunable pores for measuring concentrations of synthetic and biological nanoparticle dispersions. Biosens Bioelectron. 31 (1), 17-25 (2012).
- Roberts, G. S., Kozak, D., Anderson, W., Broom, M. F., Vogel, R., Trau, M. Tunable nano/micropores for particle detection and discrimination: scanning ion occlusion spectroscopy. Small. 6 (23), 2653-2658 (2010).
- Vogel, R., et al. Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor. Anal Chem. 83 (9), 3499-3506 (2011).
- Booth, M. A., Vogel, R., Curran, J. M., Harbison, S., Travas-Sejdic, J. Detection of target-probe oligonucleotide hybridization using synthetic nanopore resistive pulse sensing. Biosens Bioelectron. 45, 136-140 (2013).
- Kozak, D., Anderson, W., Vogel, R., Chen, S. Simultaneous size and ζ-potential measurements of individual nanoparticles in dispersion using size-tunable pore sensors. ACS Nano. 6 (8), 6990-6997 (2012).
- Kozak, D., Anderson, W., Vogel, R., Trau, M. Advances in Resistive Pulse Sensors: Devices bridging the void between molecular and microscopic detection. Nano Today. 6 (5), 531-545 (2011).
- Weatherall, E., Willmott, G. R. Applications of tunable resistive pulse sensing. Analyst. 140, 3318-3334 (2015).
- Willmott, G. R., et al. Use of tunable nanopore blockade rates to investigate colloidal dispersions. J Phys Condens Matter. 22 (45), 454116 (2010).
- Blundell, E. L. C. J., Mayne, L. J., Billinge, E. R., Platt, M. Emergence of tunable resistive pulse sensing as a biosensor. Anal Methods. 7, 7055-7066 (2015).
- Platt, M., Willmott, G. R., Lee, G. U. Resistive Pulse Sensing of Analyte-Induced Multicomponent Rod Aggregation Using Tunable Pores. Small. 8 (15), 2436-2444 (2012).
- Vogel, R., Anderson, W., Eldridge, J., Glossop, B., Willmott, G. A variable pressure method for characterizing nanoparticle surface charge using pore sensors. Anal Chem. 84 (7), 3125-3131 (2012).
- Blundell, E. L. C. J., Vogel, R., Platt, M. Particle-by-Particle Charge Analysis of DNA-Modified Nanoparticles Using Tunable Resistive Pulse Sensing. Langmuir. 32 (4), (2016).
- Hunter, R. J. . Zeta Potential in Colloid Science: Principles and Applications. , (1981).
- Arjmandi, N., Van Roy, W., Lagae, L., Borghs, G. Measuring the electric charge and zeta potential of nanometer-sized objects using pyramidal-shaped nanopores. Anal Chem. 84 (20), 8490-8496 (2012).
- Bacri, L., et al. Dynamics of colloids in single solid-state nanopores. J Phys Chem B. 115 (12), 2890-2898 (2011).
- Cabello-Aguilar, S., et al. Dynamics of polymer nanoparticles through a single artificial nanopore with a high-aspect-ratio. Soft Matter. 10 (42), 8413-8419 (2014).
- Billinge, E. R., Muzard, J., Platt, M. Tunable resistive pulse sensing as a tool to monitor analyte induced particle aggregation. Nanomater Nanosci. 1 (1), 11 (2013).
- Li, J., Fan, C., Pei, H., Shi, J., Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv Mater. 25 (32), 4386-4396 (2013).
- Billinge, E. R., Broom, M., Platt, M. Monitoring aptamer-protein interactions using tunable resistive pulse sensing. Anal Chem. 86 (2), 1030-1037 (2014).
- Gold, L., et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS One. 5 (12), e15004 (2010).
- Park, S. -. J., Taton, T. A., Mirkin, C. A. Array-Based Electrical Detection of DNA with Nanoparticle Probes. Science. 295 (5559), 1503-1506 (2002).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved