A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Analyzing the Size, Shape, and Directionality of Networks of Coupled Astrocytes
In This Article
Summary
Here we present a protocol to assess the organization of astrocytic networks. The described method minimizes bias to provide descriptive measures of these networks such as cell count, size, area, and position within a nucleus. Anisotropy is assessed with a vectorial analysis.
Abstract
It has become increasingly clear that astrocytes modulate neuronal function not only at the synaptic and single-cell levels, but also at the network level. Astrocytes are strongly connected to each other through gap junctions and coupling through these junctions is dynamic and highly regulated. An emerging concept is that astrocytic functions are specialized and adapted to the functions of the neuronal circuit with which they are associated. Therefore, methods to measure various parameters of astrocytic networks are needed to better describe the rules governing their communication and coupling and to further understand their functions.
Here, using the image analysis software (e.g., ImageJFIJI), we describe a method to analyze confocal images of astrocytic networks revealed by dye-coupling. These methods allow for 1) an automated and unbiased detection of labeled cells, 2) calculation of the size of the network, 3) computation of the preferential orientation of dye spread within the network, and 4) repositioning of the network within the area of interest.
This analysis can be used to characterize astrocytic networks of a particular area, compare networks of different areas associated to different functions, or compare networks obtained under different conditions that have different effects on coupling. These observations may lead to important functional considerations. For instance, we analyze the astrocytic networks of a trigeminal nucleus, where we have previously shown that astrocytic coupling is essential for the ability of neurons to switch their firing patterns from tonic to rhythmic bursting1. By measuring the size, confinement, and preferential orientation of astrocytic networks in this nucleus, we can build hypotheses about functional domains that they circumscribe. Several studies suggest that several other brain areas, including the barrel cortex, lateral superior olive, olfactory glomeruli, and sensory nuclei in the thalamus and visual cortex, to name a few, may benefit from a similar analysis.
Introduction
Many studies have described how the neuron-astrocyte dialogue at a sub-cellular or synaptic level can have implications in neuronal functions and synaptic transmission. It is well established that astrocytes are sensitive to surrounding neuronal activity; in fact, they have receptors for many neurotransmitters including glutamate, GABA, acetylcholine, and ATP (see previously published reviews2,3,4). In return, astrocytic processes ensheath synaptic elements and influence neuronal activity both there and at extrasynaptic sites by regulating extracellular ionic homeostasis and releasing several factors or transmitters such as glutamate, D-serine, and ATP5,6,7.
The idea that astrocytes can also modulate neuronal function at the network level has emerged, with evidence that astrocytic coupling is spatially regulated and corresponds to neuronal segmentation in areas characterized by a clear anatomical compartmentalization (like areas with sensory representations), indicating that astrocytes will couple to other astrocytes serving the same function rather than just those that are close by. In the lateral superior olive, for instance, most astrocytic networks are oriented orthogonally to the tonotopic axis8, whereas in the barrel cortex or olfactoty glomeruli, communication between astrocytes is much stronger within barrels or glomeruli and weaker between adjacent ones9,10. In both cases, the astrocytic networks are oriented towards the center of the glomerule or barrel9,10.
We recently showed that astrocytic activity modulates neuronal firing by decreasing the concentration of extracellular Ca2+ ([Ca2+]e), presumably through the release of S100β, a Ca2+-binding protein11. This effect, which was demonstrated in a population of trigeminal rhythmogenic neurons in the dorsal part of the trigeminal main sensory nucleus (NVsnpr, thought to play an important role in the generation of masticatory movements), results from the fact that rhythmic firing in these neurons depends on a persistent Na+ current that is promoted by decreases of [Ca2+]e11,12. Rhythmic firing in these neurons can be elicited "physiologically" by stimulation of their inputs or artificial decrease of [Ca2+]e. We further showed that astrocytic coupling was required for neuronal rhythmic firing1. This raised the possibility that astrocytic networks may form circumscribed functional domains where neuronal activity can be synchronized and coordinated. To assess this hypothesis, we first needed to develop a method to rigorously document the organization of these networks within NVsnpr.
Previous studies on astrocytic networks have mostly described the extent of coupling in terms of cell number and the density and area covered. Attempts to evaluate the shape of astrocytic networks and the direction of dye-coupling were mostly performed by comparing the size of networks along two axes (x and y) in the barrel cortex9, hippocampus13,14,15, barreloid fields of the thalamus16, lateral superior olive8, olfactory glomeruli10, and cortex14. The methods described here enable unbiased counts of labeled cells in a network and an estimation of the area they cover. We also developed tools to define the preferred orientation of coupling within a network and to assess whether the preferred orientation is towards the center of the nucleus or in a different direction. In comparison to previously used methods, this protocol provides a means to describe the organization and orientation of astrocytic networks in structures like the dorsal trigeminal main sensory nucleus that do not have a known clear anatomical compartmentalization. In the above studies, the network orientation is described as a relationship to the shape of the structure itself which is already documented (e.g., the barreloid in the thalamus, barrels in the cortex, layers in the hippocampus and cortex, glomeruli in the olfactory bulb, etc.). In addition, vectorial analysis allows for comparisons of coupling orientations revealed under different conditions. To analyze whether these parameters changed according to the position of the network within the nucleus, we also developed a method to replace each network in reference to the boundaries of the nucleus. These tools can be easily adapted to other areas for investigating networks of coupled cells.
Protocol
All procedures abode by the Canadian Institutes of Health Research rules and were approved by the University of Montreal Animal Care and Use Committee.
1. Preparation of Rat Brain Slices
- Prepare 1 L of a sucrose-based solution (Table 1) and 1 L of standard artificial cerebral-spinal fluid (aCSF) (Table 2).
- Bubble the sucrose-based solution with a mix of 95% O2 and 5% CO2 (carbogen) for 30 min before placing it in -80 °C for about 30 min, until the solution is cold but not entirely frozen. Use this ice-cold sucrose as the cutting buffer for the slicing of the brain. Keep it on ice once it is removed from the freezer.
- Bubble the aCSF with carbogen through the entire experiment. Use this solution for slice storage and as the perfusing buffer in which the patch-clamping and biocytin-filling of astrocytes will be performed. Prepare a slice recovery holding chamber to deposit the brain slices as soon as they are cut.
Note: The slice recovery holding chamber could be custom-made and consists essentially of one small well with a mesh at the bottom suspended in a larger well that is filled with aCSF, in which a tube is inserted to bring carbogen from the bottom. - Use 15- to 21-day-old Sprague-Dawley rats without any sex- or strain-specific bias. Anesthetize the animal with isoflurane (1 mL of isoflurane in an anesthesia induction chamber). Check the depth of anesthesia by gently pinching the hind paw or tail of the animal.
- Decapitate the rat using a guillotine, cut its skull with scissors, and quickly remove the brain from the cranium with a flat spatula.
- Dip the brain in the ice-cold sucrose-based solution for about 30 s and transfer it (with the solution) to a petri dish. With a razor blade, remove the portions that are anterior and posterior to the area to be sectioned, if cutting slices in the transverse plane.
- Glue the remaining block of brain tissue on its rostral side and proceed to cut brain slices (350 µm thick) in the sucrose-based medium using a vibratome. Then, transfer the collected slices in a recovery holding chamber filled with carbogenated aCSF at room temperature (RT) until they are ready to be used (allow at least 1 hour for recovery).
Note: Recent developments in the field allow prolonged incubation up to 24 h of brain slices in in vitro conditions17.
2. Sulforhodamine 101 (SR-101) Labeling of Astrocytes
- Pre-heat a water bath to 34 °C and place two slice incubation chambers in it. Fill one of the slice incubation chambers with a solution of aCSF containing 1 μM SR-101, and fill the other only with aCSF.
- Incubate the slices in the incubation chamber containing the 1 µM SR-101 for 20 min and then transfer them into the second incubation chamber to rinse out excess SR-101 from the tissue. Let it incubate for 20 min or more at 34 °C, then keep the incubation chamber containing the slices at RT until they are needed18.
3. Astrocyte Patching and Biocytin Filling
- Select a slice and place it in the recording chamber of the microscope. To patch astrocytes, use electrodes with a resistance of 4-6MΩ when filled with a potassium-gluconate-based solution (see Table 3).
- Under visual guidance and using a micromanipulator, direct the recording electrode towards an SR-101-labeled astrocyte as shown in Figure 1. Avoid cells located at the surface of the slice since they are more likely to be damaged or have lost connections to neighboring cells.
- To prevent leakage of biocytin in the tissue, minimize the positive pressure added to the patch pipette and apply it only when it is close to the astrocyte that will be patched (0.1-0.4 mL in a syringe of 1 mL).
- Prior to patching, adjust the offset of the pipette and its capacitance. Correct for liquid junction potentials, as accurate and precise voltage commands are critical to the experiments19.
- Move the pipette close enough to the astrocyte to observe depression caused by the positive pressure. Then, remove the positive pressure and slowly apply some negative pressure. Clamp the astrocyte to -70 mV when the seal reaches 100 MΩ. Wait until the seal reaches 1-3 GΩ. Continue to apply negative pressure until breaking into the cell. Be careful while applying negative pressure, since astrocytes are very fragile.
- Assess the electrophysiological properties of the patched cell. In voltage-clamp mode, perform a whole-cell current-voltage protocol with a ramp voltage command of 600 ms duration ranging from -120 to +110 mV. In current-clamp mode, perform a step IV protocol in which the injection of 1000 ms current steps of 100 pA from -1 to 1 nA, the sampling rate of the whole-cell recordings is 10 kHz.
Note: Astrocytes show a linear current-voltage profile without any kind of rectification (as shown in Figure 1B) and no action potential firing upon membrane depolarization (Figure 1C). The resting membrane potential (RMP) should be stable and not positive to -60mV. In some brain areas, the RMP of astrocytes is more hyperpolarized than in the NVsnpr. - Allow the biocytin to diffuse within the astrocyte for 30 min while performing a step IV protocol every 5 min.
- Retract and detach the patch pipette carefully without damaging the patched astrocyte and immediately take note of the offset of the pipette before taking it out of the recording chamber. Subtract that value from the membrane potential recordings.
- Leave the brain slice to rest in the recording chamber for a minimum of 15 min (in addition to the 30 min for injection) to allow spreading of the tracer from the patched astrocyte to the entire network of coupled cells.
Note: To reveal a network of coupled cells, only a single cell should be patched in the nucleus of interest. If multiple attempts are required in order to achieve a successful patch, discard the case and try to patch on the contralateral side or in another brain slice. - Make a mark or incision in the tissue to identify which side of the slice is facing upwards, and transfer the slice first to a petri dish containing normal aCSF, then into a solution of 4% paraformaldehyde (PFA). Incubate the slice at 4 °C overnight.
CAUTION: PFA is extremely harmful. Use protective equipment in a ventilated hood. Ensure that all materials (brushes, tubes, etc.) that have been in contact with PFA do not contact the recovery holding chamber, incubation chamber, or other materials used for recording the fresh tissue.
4. Biocytin Revelation
- Revelation with fluorescent streptavidine
- Wash the brain slices 2 x 10 min in 0.1 M phosphate buffer saline (PBS) at RT. Then, reveal the biocytin by incubating the brain slices with streptavidine conjugated to a fluorophore at 1/200 dilution in PBS with 4% non-ionic detergent for 4 h at RT.
- Wash the brain slices 3 x 10 min in 0.1 M PBS at RT and mount the sections on glass slides using an aqueous mounting medium. Place the sides that were injected upwards, coverslip the slides, and seal them with nail-varnish.
- Revelation with DAB
Note: DAB (3,3-diaminobenzidine) revelation can be used when fluorescence cannot be used. In this case, only one revelation method should be employed for making comparisons.- Wash the brain slices 3 x 10 min in PBS at RT. Incubate the slices in PBS + H2O2 0.5% for 1 hour at RT.
- Rinse the slices 3 x 10 min in PBS at RT. Then, incubate the slices in an avidin-biotin complex staining solution composed of PBS and 0.1% non-ionic detergent + avidin-biotin complex peroxidase standard staining kit reagents at 1/100 for 24 hours at RT.
- Rinse the slices 3 x 10 min in PBS at RT, incubate them in a solution of PBS + DAB 0.05% for 20 min at RT, and transfer them into a solution of PBS + DAB 0.05% + H2O2 0.5%.
CAUTION: DAB is extremely harmful. Use protective equipment in a ventilated hood. The color reaction is stopped when the slices are transferred in PBS. - Rinse the slices 5 x 10 min in PBS at RT, mount the sections on glass slides (face the sides that were injected upwards) and let them dry on a slide drying bench overnight at 34 °C.
- Submerge the glass slides for 1 min in several alcohol baths at 70%, 95%, and 100%, and end with a bath of xylene for 1 min.
- Mount the glass slides using a toluene-based synthetic resin mounting medium. Place coverslips on the slides and seal them with nail varnish.
5. Network Imaging
- Visualize the astrocytic networks using a scanning confocal microscope equipped with 20X and 4X objectives (or an appropriate objective to visualize the entire nucleus where the network is located) and a laser to detect the fluorophore (in this case, Alexa-594 is used).
- Use the 20X magnification to make a z-stack of the network of labeled cells. Use a resolution of 800 x 800 pixels and scan speed of 12.5 μs/pixel.
Note: In order to image the whole network, multiple stacks are usually required, and the number of stacks needs to be adjusted for each network. The resolution and scan speed can be changed, but make sure to use the same settings to image all data. - Use the 4X magnification to take images of the network and region of interest.
Note: The 4X imaging is used to determine the network position within the nucleus of interest. Always image the same field in transmitted light. This image will be useful if you cannot determine the border of the nucleus in the confocal fluorescent image.
6. Image Analysis
- Data preparation
- Use the software ImageJFIJI (download it at https://fiji.sc/). Open the file and click OK in the “Bio-Formats Import Options” window.
- To redefine a z-stack that will contain only the optical slices needed for the final z-stack (Figure 2A), click on the “Stack” knob in the toolbar (to find, first select stk | Z Project). Select “Max Intensity” in the projection type setting (Figure 2A). Save the file and name it “stack file”.
- If the imaging file contains several channels, split it to conserve only the channel with the astrocytic network imaging (Image | Color | Split Channels).
- Check the pixel settings of the image [Image | Properties | Pixel (with "1" for pixel dimension)].
- Use the subtract background tool (Process | Subtract Background) to remove the background of biocytin labelling. Use the preview function to set the rolling ball radius, which is generally set at 50 pixels (Figure 2B).
- Use the remove outliers tool (Process | Noise | Remove Outliers) if it is required following the subtract background step. Select “Bright” in the “Which Outliers” setting (Figure 2C). Use the preview function to set the radius and threshold. Be careful with this tool, since it may blur the data, as shown in Figure 2D.
Note: This tool removes the small spots caused by unspecific deposits of streptavidine (as shown by the white arrows in Figure 2B and 2C before and after treatment, respectively). - Adjust the threshold (Image | Adjust | Threshold). Select “Default” and “B&W” mode (Figure 2E). Click on “Apply”.
Note: Auto-adjusting can be used, but manual adjusting with the two slider bars is preferred. The aim of this step is to reduce noise to the maximum without losing any labeled cells. - Convert the image into a binary image with the binary process tool (Process | Binary | Make Binary) as shown in Figure 2F. Save the file as a TIFF file and name it “binary file”.
- Cell counting
- Check the setting of the measure function (Analyze | Set measurement). Select the “Centroid” option.
- Use the “Analyze Particles” tool on the “binary file” (Figure 3A) produced in the previous step (Analyze | Analyze Particle) (Figure 3C on the left). Select “Outlines” in the “Show” setting (Figure 3C). This generates a new file that shows the result of the detection (Figure 3B).
- Play with the parameters: size (to detect only cells, use values between 30 and 6000) and circularity (to determine an interval between 0 to 1, in which “1” defines a perfect circle and “0” a random shape) to refine the detection (Figure 3C, left portion). Run the detection by clicking “OK”.
Note: Two tables will appear following the detection: 1) a table titled “Summary” that provides the number of detected cells, and 2) a table titled “Results” (Figure 3C, right portion) that provides the x- and y-coordinates of each cell. - Copy the values and paste them into a spreadsheet application. Save this table under the name “detection table”. A file with a plot of detected cells will also appear (Figure 3B). Save this file as a TIFF file under the name “detection file”.
- If a group of 2 or more labeled cells in the network are detected as a single cell with the analyze particles tool because they are too close to each other, use the watershed tool (Process | Binary | Watershed) on the binary image before applying the analyze particles tool, and redo the analyze particles steps.
Note: The watershed tool creates a delimitation of 1 pixel wide between extremely close cells. A cell counter plug-in can be used when labeling is unequivocal and can be conducted manually.
- Measuring astrocytic network area
- To measure the surface area of the networks, use the detection file using Image J.
- Use the polygon selection tool (left click on the button in the tool bar to select it) to trace a polygon that connects all the cells located in the external periphery of the network (Figure 4A). Left click to begin tracing the polygon, and right click to close it.
Note: This polygon will be defined as a region of interest (ROI) and its surface will be measured to determine the surface area of the network. - Open the set measurement window (Analyze | Set Measurement) and select the “Area” option. Open the ROI Manager (Analyze | Tools | ROI Manager) (Figure 4B). Then, add the traced polygon in the ROI Manager by clicking ‘’Add’’ (Figure 4B) and run the measurement by clicking ‘’Measure’’ in the ROI Manager.
Note: The area measurement will appear in a table and be expressed in pixels. Do not forget to convert this value with the conversion factor for the microscope used to obtain a value in μm2.
- Determination of the main direction vector
- Determination of the patched cell
- Open the stack file in ImageJFIJI and identify the patched cell in the stack file based on its stronger labeling intensity (Figure 4C). Then, open the file named “detection table” in a spreadsheet application and find the number associated to the patched cell and its corresponding coordinates.
- If unable to accurately determine the patched cell, surround the area where the deposit of biocytin is denser in the imaged network of coupled cells, using the Polygon tool in ImageJFIJI, and refer to this position as that of the patched cell (Figure 4D).
- Use the ROI Manager (Analyze | Tools | ROI Manager). Then, draw a ROI at the patched cell location and add it to the ROI Manager (see Figure 4B).
- Set a measurement (Analyze | Set Measurement) and select the “Centroid” option.
- In the ROI Manager, click on “Measure” to obtain the coordinates of the centroid of the traced area. Use these coordinates as the reference point for this specific network.
- Referential translation
- Calculate the coordinates of each cell in reference to the patched astrocyte by using the following formula:

With: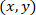 as the coordinates for a given cell;
as the coordinates for a given cell; 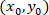 as the coordinates of the patched cell (or the referential point of the network); and
as the coordinates of the patched cell (or the referential point of the network); and 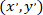 as the coordinates for a given cell in the new referential.
as the coordinates for a given cell in the new referential.
Note: Expressing the coordinates of each cell in reference to the patched astrocyte is an important step in calculating vectors from the patched astrocyte. Be careful when using ImageJFIJI that the referential for any image is located in the top-left corner of the image.
- Calculate the coordinates of each cell in reference to the patched astrocyte by using the following formula:
- Determination of the main vector of preferential orientation
- Calculate the coordinates of the main vector of preferential orientation with the following formula:
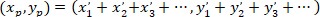
With: (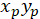 ) as the coordinates of the main vector of preferential orientation; and
) as the coordinates of the main vector of preferential orientation; and 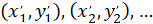 as the coordinates of each cell of the network obtained with the patched cell as referential.
as the coordinates of each cell of the network obtained with the patched cell as referential.
Note: For each cell in the network, a vector is determined relative to the coordinates of the patched astrocyte. The main vector of preferential orientation of the network is the sum of all these vectors. - Divide the length of the main vector (provided by the coordinates obtained from using the above formula) by the number of cells in the network minus one (since the coordinates of the patched cell are not included), to normalize the values and enable comparisons between networks. A schematic view of this analysis is presented in Figure 5.
- Calculate the coordinates of the main vector of preferential orientation with the following formula:
- Determination of the patched cell
- Placement of analyzed networks in the nucleus of interest
- Alignment of the 20X and 4X images
- To determine the position of each network in the nucleus of interest (NVsnpr), use the 4X image. Open the 4X image with a vector image editor.
- Select the 4X image and modify its size by multiplying it by 5. The dimension window is located in the right part of the top horizontal toolbar. For instance, for a 4X image that has been sampled at 800 x 800 pixels, change the sampling resolution to 4000 x 4000 pixels (to set the working unit in pixels, go to “Document Setup” under the file tab: File | Document Setup). Export the file in TIFF format and name it “resized 4X”.
- Align the top-left corner of the image with the bottom-left corner of the Adobe Illustrator document.
Note: The software provides coordinates from a bottom-left corner referential point. - Open the 20X image of the network. Use the ‘’binary file’’ or ‘’detection file’’ because they are easier to align. Then, play with the opacity tool on the ‘’binary file’’ of the 20X image to align it with the re-sized 4X image.
Note: The opacity tool is in the top horizontal tool bar. - When the alignment is done, select and hold the pipette tool so the measure tool appears, and select it in the left toolbar.
- Use the measure tool to click on the top-left corner of the 20X image to get the coordinates of the 20X referential point onto the resized 4X image.
Note: These coordinates, which are referred to as the 20X referential (20XR in Figure 5), will be useful for expressing the position of each network on a schematic drawing of the nucleus.
- Normalization of the nucleus of interest
Note: To sum up the data, a normalization of the nucleus (NVsnpr) as a rectangle is used. The steps are described below.- Open the 4X resized file in ImageJFIJI, and use the polygon tool to surround the nucleus (NVsnpr). Use the image with transmitted light if the borders of the nucleus are not able to be seen; in which case, remember to resize it first.
- Open the ROI Manager and add the drawn ROI. In “Set Measurement”, select the ‘’Bounding Rectangle’’ option.
Note: The “Bounding Rectangle” option calculates the smallest rectangle around the drawn nucleus. - Click “Measure”.
Note: A table appears with BX and BY, the coordinates of the top left corner of the rectangle: ‘’Rectangle Position’’ and provides the width and height. BX and BY are the coordinates of the bounding rectangle referential which are referred to as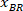 and
and 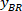 .
.
- Expression of each network position in the normalized nucleus
- Express the coordinates of each cell with the 4X referential (black square in Figure 5) using the following formula:

Where: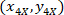 are the cell coordinates with the 4X referential;
are the cell coordinates with the 4X referential; 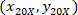 are the cell coordinates with the 20X referential (orange square in Figure 5); and
are the cell coordinates with the 20X referential (orange square in Figure 5); and 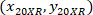 are the coordinates of the 20X referential point in the 4X image (20XR).
are the coordinates of the 20X referential point in the 4X image (20XR). - Express the cell coordinates in the bounding rectangle referential (blue in Figure 5) using the following formula:
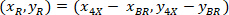
Where: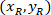 are the cell coordinates in the bounding rectangle referential;
are the cell coordinates in the bounding rectangle referential; 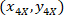 are the cell coordinates in the 4X referential; and
are the cell coordinates in the 4X referential; and  are the coordinates of the bounding rectangle referential in the 4X image.
are the coordinates of the bounding rectangle referential in the 4X image. - Transform the cell coordinates in the bounding rectangle referential to the percentage of width and height of the bounding rectangle using the following formula:
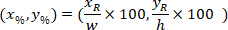
Where: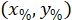 are the cell coordinates in the percentage of width and height of the bounding rectangle;
are the cell coordinates in the percentage of width and height of the bounding rectangle; 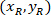 are the cell coordinates in the bounding rectangle referential;
are the cell coordinates in the bounding rectangle referential;  is the width of the bounding rectangle measured above in the protocol; and
is the width of the bounding rectangle measured above in the protocol; and 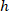 is the height of the bounding rectangle measured above in the protocol.
is the height of the bounding rectangle measured above in the protocol. - To represent all the networks on the same figure, take into account the orientation of the slice (left or right side). To standardize the data, the referential is applied on the left side of the slice. Transfer the network coordinates of the right side to the left side by applying the following formula only to x-coordinates:
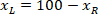
Where: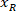 is the x-coordinate on the right side of the slice; and
is the x-coordinate on the right side of the slice; and 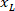 is the new x-coordinate expressed in the referential on the left side of the slice.
is the new x-coordinate expressed in the referential on the left side of the slice.
Note: Alternatively, mirror the picture in ImageJFIJI before the analysis (Image | Transform | Flip Horizontally). - To express the coordinates of the main vector of preferential orientation, follow the same steps with the cell coordinates (steps 6.5.3.1 to 6.5.3.4).
Note: The expression of the coordinates in percentages allows a compilation of the data as a single plot in which the nucleus (NVsnpr) is designed as a rectangle.
- Express the coordinates of each cell with the 4X referential (black square in Figure 5) using the following formula:
- Alignment of the 20X and 4X images
- Study of the angular difference of the main vector of preferential direction
Note: The angular difference of an astrocytic network is used to determine whether its preferential orientation is towards the center of the nucleus of interest. To calculate the angular difference (α in Figure 5), which is the angle between the main vector of preferential direction of the network (PD, red line in Figure 5) and the line connecting P to C, apply the Al-Kashi Theorem into the triangle PDC (see inset of Figure 5 and Supplementary Methods).- First, calculate the different lengths using the application of the Pythagorean Theorem into a Cartesian referential using the following equation:
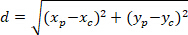
Where: the coordinates of P and C are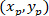 and
and  , respectively, in the bounding rectangle referential (calculated above).
, respectively, in the bounding rectangle referential (calculated above). - Determine the angular difference in radians using the following formula:
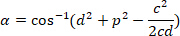
- Convert the angular difference in degrees (e.g., with the DEGREES function in Excel software).
- Compile all the angular differences by plotting them in vertical bar charts (Figure 7C and 7D) and determine if there is a preferential orientation of astrocytic networks.
- First, calculate the different lengths using the application of the Pythagorean Theorem into a Cartesian referential using the following equation:
Results
Coupling between cells in the brain is not static but rather dynamically regulated by many factors. The methods described were developed to analyze astrocytic networks revealed under different conditions and to understand their organization in NVsnpr. These results have been already published1. We performed biocytin filling of single astrocytes in the dorsal part of the NVsnpr in three different conditions: at rest (in control conditions in the absence of any stimu...
Discussion
A number of electrophysiological methods exist to assess functional coupling between astrocytes23,24. However, these methods do not provide information about the anatomical arrangement of astrocytic networks. A number of studies have already shown that "dye- or tracer-coupling", as done here, occurs only in a fraction of coupled cells that are detected by electrophysiological methods25,26,
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work is funded by the Canadian Institutes of Health Research, Grant/Award Number: 14392.
Materials
| Name | Company | Catalog Number | Comments |
| NaCl | Fisher Chemicals | S671-3 | |
| KCl | Fisher Chemicals | P217-500 | |
| KH2PO4 | Fisher Chemicals | P285-500 | |
| MgSO4 | Fisher Chemicals | M65-500 | |
| NaHCO3 | Fisher Chemicals | S233-500 | |
| C6H12O6 Dextrose anhydrous | Fisher Chemicals | D16-500 | |
| CaCl2 dihydrated | Sigma | C70-500 | |
| Sucrose | Sigma | S9378 | |
| D-gluconic acid potassium salt | Sigma | G45001 | |
| MgCl2 anhydrous | Sigma | M8266 | |
| HEPES | Sigma | H3375 | |
| EGTA | Sigma | E4378 | |
| ATPTris Salt | Sigma | A9062 | |
| GTPTris Salt | Sigma | G9002 | |
| Biocytin | Sigma | B4261 | |
| Carbenoxolone disodium salt | Sigma | C4790 | |
| avidin-biotin complex : ABC kit | Vestor laboratories | PK-4000 | |
| Streptavidine-alexa 594 | Molecular Probes | S11227 | |
| Triton | Fisher Chemicals | BP151-500 | |
| Xylene | Fisher Chemicals | X5-1 | |
| Aqueous mounting medium 1 : Fluoromount-G | SouthernBiotech | 0100-01 | |
| Toluen-based synthetic resin mounting medium : Permount | Fisher Chemicals | SP15-100 | |
| Slide Drying Bench | Fisherbrand | 11-474-470 | |
| Vibratome | Leica | VT 1000S | |
| Microscope cover glass | Fisherbrand | 12-544A | |
| Microscope slide ColorFrost | Fisherbrand | 12-550-413 | |
| PFA | Fisherchemicals | 04042-500 | |
| Olympus FluoView FV 1000 Confocal microscope | Olympus | ||
| 40X water-immersion lens | Olympus | LUMPLFLN40XW | |
| 20X water-immersion lens | Olympus | XLUMPLFL20XW | |
| 4X water-immersion lens | Olympus | XLFLUOR4X/340 | |
| Micropipette puller | Sutter Instrument | P97 | |
| Micromanipulator | Sutter Instrument | MP 225 | |
| Camera CCD | Sony | CX-ST50 | |
| Black and white monitor | Sony | SSM-125 | |
| Digidata | Molecular devices | 1322A | |
| Patch Clamp amplifier | Axon instrument | Mulitclamp 700A | |
| Electrophysiology acquisition software | Molecular devices | pClamp 8 | |
| Electrophysiology analysis software | Molecular devices | Clampfit 8 | |
| Imaging analysis software | ImageJFIJI | Open source software. FIJI version including plug in package. | |
| Vector image editor | Adobe | Illustrator CS4 | |
| Spreadsheet application | Microsoft Office | Excel 2010 |
References
- Condamine, S., Lavoie, R., Verdier, D., Kolta, A. Functional rhythmogenic domains defined by astrocytic networks in the trigeminal main sensory nucleus. Glia. 66 (2), 311-326 (2018).
- Verkhratsky, A., Orkand, R. K., Kettenmann, H. Glial calcium: homeostasis and signaling function. Physiological Review. 78 (1), 99-141 (1998).
- Christensen, R. K., Petersen, A. V., Perrier, J. F. How do glial cells contribute to motor control?. Current Pharmaceutical Design. 19 (24), 4385-4399 (2013).
- Verkhratsky, A., Steinhauser, C. Ion channels in glial cells. Brain Research Review. 32 (2-3), 380-412 (2000).
- Harada, K., Kamiya, T., Tsuboi, T. Gliotransmitter Release from Astrocytes: Functional, Developmental, and Pathological Implications in the Brain. Frontiers Neuroscience. 9, 499 (2015).
- Montero, T. D., Orellana, J. A. Hemichannels: new pathways for gliotransmitter release. Neuroscience. 286, 45-59 (2015).
- Araque, A., et al. Gliotransmitters travel in time and space. Neuron. 81 (4), 728-739 (2014).
- Augustin, V., et al. Functional anisotropic panglial networks in the lateral superior olive. Glia. 64 (11), 1892-1911 (2016).
- Houades, V., Koulakoff, A., Ezan, P., Seif, I., Giaume, C. Gap junction-mediated astrocytic networks in the mouse barrel cortex. Journal of Neuroscience. 28 (20), 5207-5217 (2008).
- Roux, L., Benchenane, K., Rothstein, J. D., Bonvento, G., Giaume, C. Plasticity of astroglial networks in olfactory glomeruli. Proceedings of the National Academy of Science of the United State of America. 108 (45), 18442-18446 (2011).
- Morquette, P., et al. An astrocyte-dependent mechanism for neuronal rhythmogenesis. Nature Neuroscience. 18 (6), 844-854 (2015).
- Brocard, F., Verdier, D., Arsenault, I., Lund, J. P., Kolta, A. Emergence of intrinsic bursting in trigeminal sensory neurons parallels the acquisition of mastication in weanling rats. Journal of Neurophysiology. 96 (5), 2410-2424 (2006).
- Anders, S., et al. Spatial properties of astrocyte gap junction coupling in the rat hippocampus. Philosophical Transactions of the Royal Society of London. Series B, Biological Science. 369 (1654), (2014).
- Houades, V., et al. Shapes of astrocyte networks in the juvenile brain. Neuron Glia Biology. 2 (1), 3-14 (2006).
- Rouach, N., Koulakoff, A., Abudara, V., Willecke, K., Giaume, C. Astroglial metabolic networks sustain hippocampal synaptic transmission. Science. 322 (5907), 1551-1555 (2008).
- Claus, L., et al. Barreloid Borders and Neuronal Activity Shape Panglial Gap Junction-Coupled Networks in the Mouse Thalamus. Cerebral Cortex. 28 (1), 213-222 (2018).
- Cameron, M. A., et al. Prolonged Incubation of Acute Neuronal Tissue for Electrophysiology and Calcium-imaging. Journal of Visualized Experiments. (120), (2017).
- Kafitz, K. W., Meier, S. D., Stephan, J., Rose, C. R. Developmental profile and properties of sulforhodamine 101--Labeled glial cells in acute brain slices of rat hippocampus. Journal of Neuroscience Methods. 169 (1), 84-92 (2008).
- Neher, E. Correction for liquid junction potentials in patch clamp experiments. Methods in Enzymology. , 123-131 (1992).
- Giaume, C., Leybaert, L., Naus, C. C., Saez, J. C. Connexin and pannexin hemichannels in brain glial cells: properties, pharmacology, and roles. Frontiers in Pharmacology. 4, 88 (2013).
- Torres, A., et al. Extracellular Ca(2)(+) acts as a mediator of communication from neurons to glia. Science Signaling. 5 (208), ra8 (2012).
- Ye, Z. C., Wyeth, M. S., Baltan-Tekkok, S., Ransom, B. R. Functional hemichannels in astrocytes: a novel mechanism of glutamate release. Journal of Neuroscience. 23 (9), 3588-3596 (2003).
- Ma, B., et al. Gap junction coupling confers isopotentiality on astrocyte syncytium. Glia. 64 (2), 214-226 (2016).
- Meme, W., Vandecasteele, M., Giaume, C., Venance, L. Electrical coupling between hippocampal astrocytes in rat brain slices. Neuroscience Research. 63 (4), 236-243 (2009).
- Ransom, B. R., Kettenmann, H. Electrical coupling, without dye coupling, between mammalian astrocytes and oligodendrocytes in cell culture. Glia. 3 (4), 258-266 (1990).
- Audesirk, G., Audesirk, T., Bowsher, P. Variability and frequent failure of lucifer yellow to pass between two electrically coupled neurons in Lymnaea stagnalis. Journal of Neurobiology. 13 (4), 369-375 (1982).
- Ewadinger, N., Syed, N., Lukowiak, K., Bulloch, A. Differential Tracer Coupling between Pairs of Identified Neurones of the Mollusc Lymnaea Stagnalis. Journal of Experimental Biology. 192 (1), 291-297 (1994).
- Griemsmann, S., et al. Characterization of Panglial Gap Junction Networks in the Thalamus, Neocortex, and Hippocampus Reveals a Unique Population of Glial Cells. Cerebral Cortex. 25 (10), 3420-3433 (2015).
- Kuwajima, T., et al. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 140 (6), 1364-1368 (2013).
- Gourine, A. V., et al. Astrocytes control breathing through pH-dependent release of ATP. Science. 329 (5991), 571-575 (2010).
- Forsberg, D., Ringstedt, T., Herlenius, E. Astrocytes release prostaglandin E2 to modify respiratory network activity. eLife. 6, (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved