A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
An Assay for Quantifying Protein-RNA Binding in Bacteria
In This Article
Summary
In this method, we quantify the binding affinity of RNA binding proteins (RBPs) to cognate and non-cognate binding sites using a simple, live, reporter assay in bacterial cells. The assay is based on repression of a reporter gene.
Abstract
In the initiation step of protein translation, the ribosome binds to the initiation region of the mRNA. Translation initiation can be blocked by binding of an RNA binding protein (RBP) to the initiation region of the mRNA, which interferes with ribosome binding. In the presented method, we utilize this blocking phenomenon to quantify the binding affinity of RBPs to their cognate and non-cognate binding sites. To do this, we insert a test binding site in the initiation region of a reporter mRNA and induce the expression of the test RBP. In the case of RBP-RNA binding, we observed a sigmoidal repression of the reporter expression as a function of RBP concentration. In the case of no-affinity or very low affinity between binding site and RBP, no significant repression was observed. The method is carried out in live bacterial cells, and does not require expensive or sophisticated machinery. It is useful for quantifying and comparing between the binding affinities of different RBPs that are functional in bacteria to a set of designed binding sites. This method may be inappropriate for binding sites with high structural complexity. This is due to the possibility of repression of ribosomal initiation by complex mRNA structure in the absence of RBP, which would result in lower basal reporter gene expression, and thus less-observable reporter repression upon RBP binding.
Introduction
RNA binding protein (RBP)-based post-transcriptional regulation, specifically characterization of the interaction between RBPs and RNA, has been studied extensively in recent decades. There are multiple examples of translational down-regulation in bacteria originating from RBPs inhibiting, or directly competing with, ribosome binding1,2,3. In the field of synthetic biology, RBP-RNA interactions are emerging as a significant tool for the design of transcription-based genetic circuits4,5. Therefore, there is an increase in demand for characterization of such RBP-RNA interactions in a cellular context.
The most common methods for studying protein-RNA interactions are the electrophoretic mobility shift assay (EMSA)6, which is limited to in vitro settings, and various pull-down assays7, including the CLIP method8,9. While such methods enable the discovery of de novo RNA binding sites, they suffer from drawbacks such as labor-intensive protocols and expensive deep sequencing reactions and may require a specific antibody for RBP pull-down. Due to the susceptible nature of RNA to its environment, many factors can affect RBP-RNA interactions, emphasizing the importance of interrogating RBP-RNA binding in the cellular context. For example, we and others have demonstrated significant differences between RNA structures in vivo and in vitro10,11.
Based on the approach of a previous study12, we recently demonstrated10 that when placing pre-designed binding sites for the capsid RBPs from the bacteriophages GA13, MS214, PP715, and Qβ16 in the translation initiation region of a reporter mRNA, reporter expression is strongly repressed. We present a relatively simple and quantitative method, based on this repression phenomenon, to measure the affinity between RBPs and their corresponding RNA binding sites in vivo.
Protocol
1. System Preparation
- Design of binding-site plasmids
- Design the binding site cassette as depicted in Figure 1. Each minigene contains the following parts (5' to 3'): Eagl restriction site, ∼40 bases of the 5' end of the kanamycin (Kan) resistance gene, pLac-Ara promoter, ribosome binding site (RBS), AUG of the mCherry gene, a spacer (δ), an RBP binding site, 80 bases of the 5' end of the mCherry gene, and an ApaLI restriction site.
NOTE: To increase the success rate of the assay, design three binding-site cassettes for each binding site, with spacers consisting of at least one, two, and three bases. See Representative Results section for further guidelines.
- Design the binding site cassette as depicted in Figure 1. Each minigene contains the following parts (5' to 3'): Eagl restriction site, ∼40 bases of the 5' end of the kanamycin (Kan) resistance gene, pLac-Ara promoter, ribosome binding site (RBS), AUG of the mCherry gene, a spacer (δ), an RBP binding site, 80 bases of the 5' end of the mCherry gene, and an ApaLI restriction site.
- Cloning of binding site plasmids
- Order the binding-site cassettes as double-stranded DNA (dsDNA) minigenes. Each minigene is ∼500 bp long and contains an Eagl restriction site and an ApaLI restriction site at the 5' and 3' ends, respectively (see step 1.1.1).
NOTE: In this experiment, mini-genes with half of the kanamycin gene were ordered to facilitate screening for positive colonies. However, Gibson assembly17 is also suitable here, in which case the binding site can be ordered as two shorter complementary single-stranded DNA oligos. - Double-digest both the mini-genes and the target vector with Eagl-HF and ApaLI by the restriction protocol18, and column purify19.
- Ligate the digested minigenes to the binding-site backbone containing the rest of the mCherry reporter gene, terminator, and a kanamycin resistance gene20.
- Transform the ligation solution into Escherichia coli TOP10 cells21.
- Identify positive transformants via Sanger sequencing.
- Design a primer 100 bases upstream to the region of interest (see Table 1 for primer sequences).
- Miniprep a few bacterial colonies22.
- Prepare 5 µL of a 5 mM solution of the primer and 10 µL of the DNA at 80 ng/µL concentration.
- Send the two solution to a convenient facility for Sanger sequencing23.
- Store purified plasmids at -20 °C, and bacterial strains as glycerol stocks24, both in the 96-well format. DNA will then be used for transformation into E. coli TOP10 cells containing one of four fusion-RBP plasmids (see step 1.3.5).
- Order the binding-site cassettes as double-stranded DNA (dsDNA) minigenes. Each minigene is ∼500 bp long and contains an Eagl restriction site and an ApaLI restriction site at the 5' and 3' ends, respectively (see step 1.1.1).
- Design and construction of the RBP plasmid
NOTE: Amino acid and nucleotide sequences of the coat proteins used in this study are listed in Table 2.- Order the required RBP sequence lacking a stop codon as a custom-ordered dsDNA minigene lacking a stop codon with restriction sites at the ends (Figure 1).
- Clone the tested RBP lacking a stop codon immediately downstream of an inducible promoter and upstream of a fluorescent protein lacking a start codon (Figure 1), similar to steps 1.2.2-1.2.4. Make sure that the RBP plasmid contains a different antibiotic resistance gene than the binding-site plasmid.
- Identify positive transformants via Sanger sequencing, similar to step 1.2.5 (see Table 1 for primer sequences).
- Choose one positive transformant and make it chemically-competent25. Store as glycerol purified plasmids at -20 °C and glycerol stocks of bacterial strains24 at -80 °C in 96-well plates.
- Transform the binding-site plasmids (from step 1.2.6) stored in 96-well plates into chemically-competent bacterial cells already containing an RBP-mCerulean plasmid21. To save time, instead of plating the cells on Petri dishes, plate them using an 8-channel pipettor on 8-lane plates containing Luria-Bertani (LB)26 agar with relevant antibiotics (Kan and Amp). Colonies should appear in 16 h.
- Select a single colony for each double transformant and grow overnight in LB medium with the relevant antibiotics (Kan and Amp) and store as glycerol stocks24 at -80 °C in 96-well plates.
2. Experiment Setup
NOTE: The protocol presented here was performed using a liquid-handling robotic system in combination with an incubator and a plate reader. Each measurement was carried out for 24 inducer concentrations, with two duplicates for each strain + inducer combination. Using this robotic system, data for 16 strains per day with 24 inducer concentrations was collected. However, if such a device is unavailable, or if fewer experiments are necessary, these can easily be done by hand using an 8-channel multi-pipette and adapting the protocol accordingly. For example, preliminary results for four strains per day with 12 inducer concentrations and four time-points were acquired in this manner.
- Prepare, in advance, 1 L of bioassay buffer (BA) by mixing 0.5 g of tryptone, 0.3 mL of glycerol, 5.8 g of NaCl, 50 mL of 1 M MgSO4, 1 mL of 10x phosphate-buffered saline (PBS) buffer pH 7.4, and 950 mL of double distilled water (DDW). Autoclave or sterile filter the BA buffer.
- Grow the double-transformant strains at 37 °C and 250 rpm shaking in 1.5 mL LB with appropriate antibiotics (kanamycin at a final concentration of 25 μg/mL and ampicillin at a final concentration of 100 μg/mL), in 48-well plates, over a period of 18 h (overnight).
- In the morning, make the following preparations.
- Inducer plate. In a clean 96-well plate, prepare wells with semi-poor medium (SPM) consisting of 95% BA and 5% LB26 in the incubator at 37 °C. The number of wells corresponds to the desired number of inducer concentrations. Add C4-HSL to the wells in the inducer plate that will contain the highest inducer concentration (218 nM).
- Program the robot to serially dilute medium from each of the highest-concentration wells into 23 lower concentrations ranging from 0 to 218 nM. The volume of each inducer dilution should be sufficient for all strains (including duplicates).
- While the inducer dilutions are being prepared, warm 180 μL of SPM in the incubator at 37 °C, in 96-well plates.
- Dilute the overnight strains from step 2.2 by a factor of 100 by serial dilutions: first dilute by a factor of 10 by mixing 100 μL of bacteria with 900 μL of SPM in 48-well plates, and then dilute again by a factor of 10 by taking 20 μL from the diluted solution into 180 μL of pre-warmed SPM, in 96-well plates suitable for fluorescent measurements.
- Add the diluted inducer from the inducer plate to the 96-well plates with the diluted strains according to the final concentrations.
- Shake the 96-well plates at 37 °C for 6 h, while taking measurements of optical density at 595 nm (OD595), mCherry (560 nm/612 nm) and mCerulean (460 nm/510 nm) fluorescence via a plate reader every 30 min. For normalization purposes, measure growth of SMP with no cells added.
3. Preliminary Results Analysis
- For each day of experiment, choose a time interval of logarithmic growth according to the measured growth curves, between the linear growth phase and the stationary (T0, Tfinal). Take approximately 6−8 time points, while discarding the first and last measurements to avoid error derived from inaccuracy of exponential growth detection (see Figure 2A, top panel).
NOTE: Discard strains that show abnormal growth curves or strains where logarithmic growth phase could not be detected and repeat the experiment. - Calculate the average normalized fluorescence of mCerulean and rate of production of mCherry, from the raw data of both mCerulean and mCherry fluorescence for each inducer concentration (Figure 2A).
- Calculate normalized mCerulean as follows:
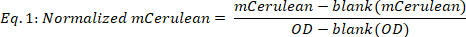
where blank(mCerulean) is the mCerulean level [a.u.] for medium only, blank(OD) is the optical density for medium only, and mCerulean and OD are the mCerulean fluorescence and optical density values, respectively. - Average mCerulean over the different time points (Figure 2B, top two panels) as follows:
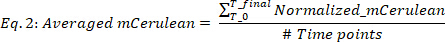
where #Time points is the number of data timepoints taken into account, T0 is the time at which the exponential growth phase begins, and Tfinal is the time at which the exponential growth phase ends. - Calculate mCherry rate of production (Figure 2B, bottom two panels) as follows:
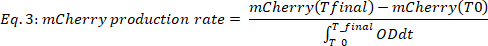
where mCherry(t) is the mCherry level [a.u.] at time t, OD is the optical density value, T0 is the time at which the exponential growth phase begins, and Tfinal is the time at which the exponential growth phase ends.
- Calculate normalized mCerulean as follows:
- Finally, plot the mCherry rate of production as a function of mCerulean, creating dose response curves as a function of RBP-mCerulean fusion fluorescence (Figure 2C). Such plots represent production of the reporter gene as a function of RBP presence in the cell.
4. Dose Response Function Fitting Routine and KRBP Extraction
- Under the assumption that the ribosome rate of translation with the RBP bound is constant, model the mCherry production rate as follows (see Figure 2D, green line):

where [x] is the normalized average mCerulean fluorescence calculated according to Eq. 2, mCherry production rate is the value calculated according to Eq. 3, KRBP is the relative binding affinity [a.u.], Kunbound is the ribosome rate of translation with the RBP unbound, n is the cooperativity factor, and C is the base fluorescence [a.u.]. C, n, Kunbound, and KRBP are found by fitting the mCherry production rate data to the model (Eq. 4). - Using data analysis software, conduct a fitting procedure on plots depicting mCherry production rate as a function of averaged mCerulean (step 3.3), and extract the fit parameters according to the formula in Eq. 4.
NOTE: Only fitting results with R2 > 0.6 are taken into account. For those fits, KRBP error is mostly in the range of 0.5% to 20% of KRBP values, for a 0.67 confidence interval, while those with higher KRBP error can be also verified by eye. - Normalize KRBP values by the respective maximal value of averaged mCerulean for each dose-response function.
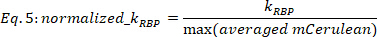
where KRBP in [a.u.] is the value extracted from the fitting procedure in Eq. 4, and max (averaged mCerulean) is the maximal averaged mCerulean signal [a.u] observed for the current strain.
NOTE: The normalization facilitates correct comparison of the regulatory effect across strains by eliminating the dependence on the particular maximal RBP expression levels.
Results
The presented method utilizes the competition between an RBP and the ribosome for binding to the mRNA molecule (Figure 1). This competition is reflected by decreasing mCherry levels as a function of increased production of RBP-mCerulean, due to increasing concentrations of inducer. In the case of increasing mCerulean fluorescence, with no significant changes in mCherry, a lack of RBP binding is deduced. Representative results for both a positive and a negativ...
Discussion
The method described in this article facilitates quantitative in vivo measurement of RBP-RNA binding affinity in E. coli cells. The protocol is relatively easy and can be conducted without the use of sophisticated machinery, and data analysis is straightforward. Moreover, the results are produced immediately, without the relatively long wait-time associated with next generation sequencing (NGS) results.
One limitation to this method is that it works only in bacterial cells. However, a...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This project received funding from the I-CORE Program of the Planning and Budgeting Committee and the Israel Science Foundation (Grant No. 152/11), Marie Curie Reintegration Grant No. PCIG11-GA- 2012-321675, and from the European Union's Horizon 2020 Research and Innovation Program under grant agreement no. 664918 - MRG-Grammar.
Materials
| Name | Company | Catalog Number | Comments |
| Ampicillin sodium salt | SIGMA | A9518 | |
| Magnesium sulfate (MgSO4) | ALFA AESAR | 33337 | |
| 48 plates | Axygen | P-5ML-48-C-S | |
| 8-lane plates | Axygen | RESMW8I | |
| 96-well plates | Axygen | P-DW-20-C | |
| 96-well plates for plate reader | Perkin Elmer | 6005029 | |
| ApaLI | NEB | R0507 | |
| Binding site sequences | Gen9 Inc. and Twist Bioscience | see Table 1 | |
| E. coli TOP10 cells | Invitrogen | C404006 | |
| Eagl-HF | NEB | R3505 | |
| Glycerol | BIO LAB | 071205 | |
| Incubator | TECAN | liconic incubator | |
| Kanamycin solfate | SIGMA | K4000 | |
| KpnI- HF | NEB | R0142 | |
| Ligase | NEB | B0202S | |
| Liquid-handling robotic system | TECAN | EVO 100, MCA 96-channel | |
| Matlab analysis software | Mathworks | ||
| Multi- pipette 8 lanes | Axygen | BR703710 | |
| N-butanoyl-L-homoserine lactone (C4-HSL) | cayman | K40982552 019 | |
| PBS buffer | Biological Industries | 020235A | |
| Platereader | TECAN | Infinite F200 PRO | |
| Q5 HotStart Polymerase | NEB | M0493 | |
| RBP seqeunces | Addgene | 27121 & 40650 | see Table 2 |
| Sodium Chloride (NaCL) | BIO LAB | 190305 | |
| SV Gel and PCR Clean-Up System | Promega | A9281 | |
| Tryptone | BD | 211705 |
References
- Cerretti, D. P., Mattheakis, L. C., Kearney, K. R., Vu, L., Nomura, M. Translational regulation of the spc operon in Escherichia coli. Identification and structural analysis of the target site for S8 repressor protein. Journal of Molecular Biology. 204 (2), 309-329 (1988).
- Babitzke, P., Baker, C. S., Romeo, T. Regulation of translation initiation by RNA binding proteins. Annual Review of Microbiology. 63, 27-44 (2009).
- Van Assche, E., Van Puyvelde, S., Vanderleyden, J., Steenackers, H. P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Frontiers in Microbiology. 6, 141 (2015).
- Chappell, J., Watters, K. E., Takahashi, M. K., Lucks, J. B. A renaissance in RNA synthetic biology: new mechanisms, applications and tools for the future. Current Opinion in Chemical Biology. 28, 47-56 (2015).
- Wagner, T. E., et al. Small-molecule-based regulation of RNA-delivered circuits in mammalian cells. Nature Chemical Biology. 14 (11), 1043 (2018).
- Bendak, K., et al. A rapid method for assessing the RNA-binding potential of a protein. Nucleic Acids Research. 40 (14), e105 (2012).
- Strein, C., Alleaume, A. -. M., Rothbauer, U., Hentze, M. W., Castello, A. A versatile assay for RNA-binding proteins in living cells. RNA. 20 (5), 721-731 (2014).
- Ule, J., Jensen, K. B., Ruggiu, M., Mele, A., Ule, A., Darnell, R. B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 302 (5648), 1212-1215 (2003).
- Lee, F. C. Y., Ule, J. Advances in CLIP Technologies for Studies of Protein-RNA Interactions. Molecular Cell. 69 (3), 354-369 (2018).
- Katz, N., et al. An in Vivo Binding Assay for RNA-Binding Proteins Based on Repression of a Reporter Gene. ACS Synthetic Biology. 7 (12), 2765-2774 (2018).
- Watters, K. E., Yu, A. M., Strobel, E. J., Settle, A. H., Lucks, J. B. Characterizing RNA structures in vitro and in vivo with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Methods. 103, 34-48 (2016).
- Saito, H., et al. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nature Chemical Biology. 6 (1), 71-78 (2010).
- Gott, J. M., Wilhelm, L. J., Uhlenbeck, O. C. RNA binding properties of the coat protein from bacteriophage GA. Nucleic Acids Research. 19 (23), 6499-6503 (1991).
- Peabody, D. S. The RNA binding site of bacteriophage MS2 coat protein. The EMBO Journal. 12 (2), 595-600 (1993).
- Lim, F., Peabody, D. S. RNA recognition site of PP7 coat protein. Nucleic Acids Research. 30 (19), 4138-4144 (2002).
- Lim, F., Spingola, M., Peabody, D. S. The RNA-binding Site of Bacteriophage Qβ Coat Protein. Journal of Biological Chemistry. 271 (50), 31839-31845 (1996).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 6 (5), 343-345 (2009).
- . Optimizing Restriction Endonuclease Reactions Available from: https://international.neb.com/tools-and-resources/usage-guidelines/optimizing-restriction-endonuclease-reactions (2018)
- . Wizard® SV Gel and PCR Clean-Up System Protocol Available from: https://worldwide.promega.com/resources/protocols/technical-bulletins/101/wizard-sv-gel-and-pcr-cleanup-system-protocol/ (2018)
- . Ligation Protocol with T4 DNA Ligase (M0202) Available from: https://international.neb.com/protocols/0001/01/01/dna-ligation-with-t4-dna-ligase-m0202 (2018)
- . Routine Cloning Using Top10 Competent Cells - US Available from: https://www.thermofisher.com/us/en/home/references/protocols/cloning/competent-cells-protocol/routine-cloning-using-top10-competent-cells.html (2018)
- . NucleoSpin Plasmid - plasmid Miniprep kit Available from: https://www.mn-net.com/ProductsBioanalysis/DNAandRNApurification/PlasmidDNApurificationeasyfastreliable/NucleoSpinPlasmidplasmidMiniprepkit/tabid/1379/language/en-US/Default.aspx (2018)
- Sanger, F., Coulson, A. R., Barrell, B. G., Smith, A. J. H., Roe, B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. Journal of Molecular Biology. 143 (2), 161-178 (1980).
- . Protocol - How to Create a Bacterial Glycerol Stock Available from: https://www.addgene.org/protocols/create-glycerol-stock/ (2018)
- . Making your own chemically competent cells Available from: https://international.neb.com/protocols/2012/06/21/making-your-own-chemically-competent-cells (2018)
- . Luria-Bertani (LB) Medium Preparation · Benchling Available from: https://benchling.com/protocols/gdD7XI0J/luria-bertani-lb-medium-preparation (2018)
- Delebecque, C. J., Silver, P. A., Lindner, A. B. Designing and using RNA scaffolds to assemble proteins in vivo. Nature Protocols. 7 (10), 1797-1807 (2012).
- Hocine, S., Raymond, P., Zenklusen, D., Chao, J. A., Singer, R. H. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nature Methods. 10 (2), 119-121 (2013).
- Espah Borujeni, A., et al. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Research. 45 (9), 5437-5448 (2017).
- Ding, Y., et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 505, (2013).
- Rouskin, S., Zubradt, M., Washietl, S., Kellis, M., Weissman, J. S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 505 (7485), 701-705 (2014).
- Lucks, J. B., et al. Multiplexed RNA structure characterization with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proceedings of the National Academy of Sciences of the United States of America. 108 (27), 11063-11068 (2011).
- Spitale, R. C., et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 519 (7544), 486 (2015).
- Watters, K. E., Abbott, T. R., Lucks, J. B. Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Research. 44 (2), e12 (2016).
- Flynn, R. A., et al. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nature Protocols. 11 (2), 273-290 (2016).
- Bernardi, A., Spahr, P. -. F. Nucleotide Sequence at the Binding Site for Coat Protein on RNA of Bacteriophage R17. Proceedings of the National Academy of Sciences of the United States of America. 69 (10), 3033-3037 (1972).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved