Method Article
Ubiquitous and Tissue-specific RNA Targeting in Drosophila Melanogaster using CRISPR/CasRx
In This Article
Summary
This article outlines a detailed protocol for using the RNA-targeting Cas13D enzyme (RfxCas13D) in flies.
Abstract
CasRx, a member of the RNA-targeting Cas13 family, is a promising new addition of the CRISPR/Cas technologies in efficient gene transcript reduction with an attractive off-target profile at both cellular and organismal levels. It is recently reported that the CRISPR/CasRx system can be used to achieve ubiquitous and tissue-specific gene transcript reduction in Drosophila melanogaster. This paper details the methods from the recent work, consisting of three parts: 1) ubiquitous in vivo endogenous RNA targeting using a two-component CasRx system; 2) ubiquitous in vivo exogenous RNA targeting using a three-component CasRx system; and 3) tissue-specific in vivo RNA targeting using a three-component CasRx system. The effects of RNA targeting observed include targeted gene specific phenotypic changes, targeted RNA transcript reduction, and occasional lethality phenotypes associated with high expression of CasRx protein and collateral activity. Overall, these results showed that the CasRx system is capable of target RNA transcript reduction at the organismal level in a programmable and efficient manner, demonstrating that in vivo transcriptome targeting, and engineering is feasible and lays the foundation for future in vivo CRISPR-based RNA targeting technologies.
Introduction
Since the advent of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technologies, much of the focus in this field has been on DNA editing, which offers transformative applications in medicine and biotechnology1. Permanent alteration of DNA sequences, however, is not always desired due to ethical considerations. In light of this, recent studies began developing CRISPR-based tools for targeting RNA and demonstrated that CRISPR technologies can indeed be used for RNA-targeting in a variety of biological systems2,3,4,5,6,7. In many of these systems tested, the current widely used approach for targeting RNA and transcript reduction is RNA interference (RNAi), which is far from perfect, often exhibiting varied efficacy and high off-target activity when used in vivo8,9,10,11,12,13,14,15,16,17. Therefore, given the status of these technologies, it is worth further exploring the potentials of CRISPR-based tools for RNA targeting.
One notable recent study reported that ribonuclease CasRx, a member of the Cas13d class, can efficiently reduce gene transcript levels in human cell culture and possesses an attractive off-target profile4. This finding led to the question of whether this new ribonuclease can maintain its efficacy and low off-target rate for RNA targeting at the organismal level. A recent study addressed this question by showing that the CasRx system can be used to achieve ubiquitous and tissue-specific gene transcript reduction in Drosophila melanogaster5.
To streamline usability of this recently published approach, this protocol details the methods from this recent work, which consists of three main parts: 1) ubiquitous in vivo RNA targeting using a two-component CasRx system; 2) ubiquitous in vivo exogenous RNA targeting using a three-component CasRx system; and 3) tissue-specificin vivo RNA targeting using a three-component CasRx system.
Guide RNAs (gRNA) targeting different target genes under the control of a ubiquitous promoter were designed and fly lines expressing these gRNA-containing constructs were generated. CasRx constructs under the control of either a ubiquitous promoter, or a conditional upstream activation sequence (UASt) promoter activatable by the GAL4 transcription factor, were also designed and fly lines harboring these CasRx-containing constructs generated. Catalytically inactive CasRx constructs, dCasRx, were designed and used as negative controls. Ubiquitous RNA targeting in flies is achieved by crossing gRNA-expressing fly lines with ubiquitously CasRx-expressing fly lines. The progeny expressing both the gRNA construct targeting a specific gene transcript and the CasRx protein has a ubiquitous reduction of targeted gene transcripts. Tissue-specific RNA targeting in flies is achieved by first crossing gRNA-expressing flies with UASt-CasRx expressing flies, obtaining transheterozygous flies carrying both gRNA and UASt-CasRx constructs. Such flies in turn are crossed with tissue-specific GAL4-expressing flies, resulting in the generation of tissue-specific CasRx expression and RNA-targeting in flies.
The programmable nature of the CasRx system offers the possibility of customization and optimization to help achieve high efficacy and low off-target activity for in vivo RNA targeting. Potential applications of CRISPR-based RNA-targeting are numerous, including replacing RNAi in the laboratory and contributing to insect vector control in the wild. Of the latter, one of the global unmet needs is the development of efficient tools to combat infections of RNA viruses transmitted via mosquitos. Many RNA viruses, such as dengue, Zika, and chikungunya virus, are transmitted via mosquitoes, affecting human health, and contributing mortality. Many proposals for engineering mosquito populations with virus resistance for disease prevention have been made; however, no current technology is able to make mosquitoes simultaneously resistant to all significant RNA viruses18,19,20,21,22,23. RNA-targeting Cas systems may provide a starting point for such a technology by enabling a programmable platform for targeting all mosquito-borne RNA viruses.
Protocol
1. Ubiquitous in vivo RNA Targeting Using a Two-Component CasRx System
- Generating Ubiq-CasRx and Ubiq-dCasRx expression vector
- Amplify the CasRx sequence using a polymerase chain reaction (PCR) with primer 1050E.C3 and 1050E.C4 and the original CasRx construct pNLS-RfxCas13d-NLS-HA (pCasRx); and amplify the dCasRx sequence using PCR with primer 1050E.C3 and 1050E.C4 and the original dCasRx construct pNLS-dRfxCas13d-NLS-HA (pdCasRx)4 (Table 1). Gel-purify the amplified CasRx and dCasRx fragments afterwards using a gel purification kit.
- Digest base vector (Addgene plasmid #112686) with restriction enzymes SwaI and PacI24. In the resulting products, use a kit to gel-purify the larger fragment, which is called the base vector backbone.
- Assemble the Ubiq-CasRx vector with the base vector backbone and the CasRx fragment using the Gibson assembly method; assemble the Ubiq-dCasRx vector with the base vector backbone and the dCasRx fragment using the Gibson assembly method25.
NOTE: The Addgene ID of Ubiq-CasRx vector (OA-1050E) is #132416, and the Addgene ID of Ubiq-dCasRx vector (OA-1050R) is #132417.
- Generating gRNA expression vector
- Design each gRNA fragment based on the following criteria: target sequence being 30 nucleotides in length; the maximal length of poly-U stretches in the target sequence being 4 base pairs; the target sequence GC content being in the range of 30% - 70%; the target sequence predicted not to form strong RNA hairpin structures; and the target sequence containing minimal predicted RNA secondary or tertiary structure5.
NOTE: This study designed each gRNA as 4 tandem sequences each 30 nucleotides long, spaced by 36 nucleotide long direct repeats, and with a 7-thymine terminator on both ends5. For the exogenous target gene, GFP, the same criteria as above was followed with an addition of a OpIE2-GFP fragment5. - Amplify the U6:3 promoter sequence using PCR with primers 1043.C1 and 1043.C23 and the Addgene plasmid #112688 (Table 1)26. Gel-purify the amplified U6:3 fragments using gel purification kit.
- Digest Addgene plasmid #112688 with restriction enzyme AscI and XbaI24. In the resulting products, use a kit to gel-purify the larger fragments, which is called the pre-base vector backbone.
- Assemble the base vector with the pre-base vector backbone and the U6:3 fragment using the Gibson assembly method25. The base vector hereafter is named OA-1043.
NOTE: The plasmid OA-1043's Addgene ID is #164586. - Synthesize target gene's gRNA fragment using external gene synthesis service.
- Digest the base vector OA-1043 with restriction enzyme PstI and NotI24. Keep the entire digestion product, which is called digested OA-1043.
- Assemble gRNA expression vector with the digested OA-1043 and target gene gRNA fragment using the Gibson assembly method25.
NOTE: Four target genes were studied: three were endogenous (white, Notch, yellow), one was exogenous (GFP). Their Addgene IDs are: #132420 (gRNAw), #132421(gRNAN), #132425 (gRNAy), and #133304 (gRNAGFP).
- Design each gRNA fragment based on the following criteria: target sequence being 30 nucleotides in length; the maximal length of poly-U stretches in the target sequence being 4 base pairs; the target sequence GC content being in the range of 30% - 70%; the target sequence predicted not to form strong RNA hairpin structures; and the target sequence containing minimal predicted RNA secondary or tertiary structure5.
- Generating transgenic flies
- Inject expression vectors into fly embryos using external fly embryo injection service and embryos from flies containing ØC31 integration sites. Rear the injected embryos at 26 °C.
NOTE: The attp40w (with integration sites on the 2nd chromosome) line was used to generate CasRx lines and 8622 (with integration sites on 3rd chromosome) line was used to generate various gRNA lines. - Keep the flies either as homozygous lines or as balanced heterozygous lines.
NOTE: Ubiq-CasRx and Ubiq-dCasRx flies were kept as heterozygous balanced lines with the CyO as the balancer. In addition, both Ubiq-CasRx and Ubiq-dCasRx vectors contain a dsRed marker. As a result, the Ubiq-CasRx and Ubiq-dCasRx flies have the following three phenotypes: dsRed-positive, curly wings, and white eyes. The gRNA-expressing flies were kept as homozygous lines. Their Bloomington Drosophila Stock Center (BDSC) fly stock numbers are: #84118 (Ubiq-CasRx), #84119 (Ubiq-dCasRx), #84124 (gRNAw), #84122 (gRNAN), #84123 (gRNAy), #84986 (gRNAGFP).
- Inject expression vectors into fly embryos using external fly embryo injection service and embryos from flies containing ØC31 integration sites. Rear the injected embryos at 26 °C.
- Fly genetics (Figure 1A)
- Collect 10 virgin adult female flies from the homozygous gRNA line and collect 5 adult male flies from the balanced heterozygous Ubiq-CasRx/CyO line. Put the collected female and male flies, which are called the parental flies, in a vial supplemented with dry yeast powder (Figure 1A).
- Repeat the previous step 3 times to generate 3 replicates. For the control group, use the Ubiq-dCasRx/CyO line while keeping everything else the same.
NOTE: For a regular glass vial of food, 0.1 g of dry yeast powder is sufficient. The BDSC’s fly food recipe is used. - Rear the vials containing the parental flies at 26 °C for 48 hours. Then remove all parental flies from every vial. Then keep the vials at 26 °C for at least 20 days.
- Observe the vials every day to see if any new adult progenies emerged from pupae of F1 generation. If so, anesthetize them with carbon dioxide by inserting a tube connected to a carbon dioxide tank inside the fly vials, then turning the flow switch on for 10 seconds.
- Once the flies become immobile, empty them from the vial onto a fly-pad, which is also connected to the carbon dioxide tank and carbon dioxide continuously flows out through the fly pad.
- Score the anesthetized flies’ phenotype and image them using color camera-connected to a fluorescent stereomicroscope. Count the numbers of progenies with different phenotypes. Use image processing software for image post-processing and compilation (Figure 2A - 2D).
NOTE: Based on Mendelian genetics, two types of flies are expected among the progenies for each cross (Figure 1A).
- RNA-Seq (Figure 2E – 2G)
- Sample collection
NOTE: Choose an appropriate sample collection method from the 3 examples below; 3 replicates for each distinct sample type are required.- Adult fly head sample collection
- Collect 10 virgin adult female flies from the homozygous gRNA line. Collect 5 adult male flies from the balanced heterozygous Ubiq-CasRx/CyO line. Put the collected female and male flies, which are the parental flies, in a vial supplemented with dry yeast powder.
- Repeat the previous step 3 times for 3 replicates. For the control group, use the Ubiq-dCasRx/CyO line while keeping everything else the same.
- Rear the vials containing the parental flies at 26 °C for 48 hours. Then remove all parental flies from every vial. Then keep the vials at 26 °C until progenies emerge from pupae.
- Collect 10 1-day old adult flies with the correct phenotype. Anesthetize the flies with carbon dioxide, then cut off the fly head and put the heads in a 1.5 mL centrifuge tube on dry ice. Store the centrifuge tube at -80 °C. Repeat this step 3 times for 3 replicates.
- 17 – 20 hour-old embryo sample collection
- Collect 8-10 virgin adult female flies from the homozygous gRNA line. Collect 4-5 adult male flies from the balanced heterozygous Ubiq-CasRx/CyO line. Put the collected female and male flies, which are the parental flies, in a vial supplemented with dry yeast powder.
- Repeat the previous step 3 times for 3 replicates. For the control group, use the Ubiq-dCasRx/CyO line while keeping everything else the same.
- Rear the vials containing the parental flies at 26 °C for 48 hours.
- Prepare one grape-juice embryo collection chamber for each replicate following this recipe: 376 mL of water, 126 mL of grape juice, 15 g of agar, and 6 g of sucrose. Put the media in a 1 L beaker and microwave it on high for 5-6 minutes while keeping a close eye on the media in the beaker to check if bubbles/foam appears. If so, stop the microwave and let the bubble/foam to settle. Continue microwaving this way until the bubble becomes clear. Do not swirl until all bubbles are clear. Finally, add 10 mL of 100% alcohol and 5 mL of acetic acid. Mix well, then pipet the media into 35 mm Petri dishes with a 25 mL serological pipet. When media solidifies in the Petri dish, it is ready for use.
- At the end of the 48-hour incubation, transfer parental flies to the grape-juice embryo collection chambers and incubated them at 26 °C for 3 h. Then remove the adult flies while keeping the freshly laid embryos on the grape-juice plates for another 17 h at 26 °C.
- After incubation, collect the 50 – 100 embryos from the grape-juice plates, clean the embryo surface by submerging them in deionized water, then transfer them to a 1.5 mL centrifuge tube on ice. Store them at -80 °C. Repeat this step 3 times for 3 replicates.
- First instar larvae sample collection
- Collect 8-10 virgin adult female flies from the homozygous gRNA line. Collect 4-5 adult male flies from the balanced heterozygous Ubiq-CasRx/CyO line. Put the collected female and male flies, which are the parental flies, in a vial supplemented with dry yeast powder. Repeat this step 3 times for 3 replicates. For the control group, use the Ubiq-dCasRx/CyO line while keeping everything else the same.
- Rear the vials containing the parental flies at 26 °C for 48 hours. Then transfer the adult flies to another new regular food vial for overnight incubation (16 h) at 26 °C. Then, remove the adult flies.
- Keep the embryo-containing vial at 26 °C for 24 h, then score the transheterozygous first instar larvae under the microscope using based distinct markers. Collect 15-30 larvae with correct phenotypes and put them into a 1.5 mL centrifuge tube and store them at -80 °C. Repeat this step 3 times for 3 replicates.
- Adult fly head sample collection
- Sequencing
- RNA extraction: use a commercially available RNA extraction kit and follow the kit’s instruction to process all samples. Then, incubate the extracted RNA samples with commercially available deoxyribonuclease and follow its instruction to remove any contaminating DNA from the samples.
- Measure RNA concentration using commercially available UV-vis spectrophotometer. Measure RNA integrity in the samples using commercially available RNA integrity assay setup.
- Construct the RNA-seq libraries using commercially available RNA library preparation kit.
- Use external sequencing service for library sequencing with the following settings: single read mode; read length: 50nt, depth: 20 million reads per library. Perform base calls with RTA 1.18.64 and then converted the data to FASTQ using bcl2fastq 1.8.4.
NOTE: Raw sequencing data can be found in the National Center for Biotechnology Information Sequencing Read Archive (submission ID: SUB6818910 [BioProject: PRJNA600654]).
- Bioinformatics
- Map reads from the sequencing data to Release 6 Drosophila melanogaster genome from the Berkeley Drosophila Genome Project (GenBank accession number: GCA_000001215.4) and the exogenous CasRx and GFP sequences using the default parameter setting of STAR aligner28 with the addition of the “-outFilterType BySJout” filter option and “-sjdbGTFfile Drosophila_melanogaster.BDGP6.22.97.gtf” gene transfer format file from ENSEMBL.
- Determine the raw transcript counts for each annotated transcript with the feature Counts35 using the “-t exon -g gene_id -M -O --fraction” options. Then, normalize raw transcript counts using total transcript counts using the “addTpmFpkmToFeatureCounts.pl” Perl script.
- Use the maximum posteriori method with the original shrinkage estimator in the DESeq2 pipeline to estimate each gene’s transcripts’ logarithmic fold change (LFC).
- Sample collection
2. Ubiquitous in vivo exogenous RNA Targeting Using a Three-Component CasRx System
- Generating exogenous target ubiquitous expression vector
- PCR amplify the Ubiq promoter fragment using primers 1052B.C1 and 1052B.C2 and the Addgene plasmid #11268626. Then, PCR amplify the T2A-eGFP fragment amplified from Addgene plasmid #112686 with primers 908A.1 and 908A.2 (Table 1)26. Then, PCR amplify the Ubiq promoter fragment as reversed sequence using Addgene plasmid #112686 with primers 908A.3 and 908A.4 (Table 1)26. Gel-purify the Ubiq promoter fragment, the T2A-eGFP fragment, and the reversed Ubiq promoter fragment using gel purification kit.
- Order a custom firefly luciferase coding sequence and a custom fragment containing a p10 3’UTR fragment, reversed renilla luciferase followed by an SV40 3’UTR fragment.
- Digest Addgene plasmid #112688 with restriction enzyme AscI and XbaI24. In the resulting products, gel-purify the larger fragments using gel purification kit, which is called the base vector backbone.
- Use the Gibson assembly method to assemble base vector with the base vector backbone and the following fragments: Ubiq promoter fragment, the T2A-eGFP fragment, the reversed Ubiq promoter fragment, the firefly luciferase coding sequence, and the reversed renilla luciferase followed by an SV40 3’UTR fragment25.
NOTE: The Addgene ID of the resulting dual-luciferase expression vector (OA-1052B) is #132426.
- Generating gRNA expression vector
- PCR amplify the U6:3 promoter sequence using primers 1043.C1 and 1043.C23 and the Addgene plasmid #112688 (Table 1)26. Gel-purify the amplified U6:3 fragments using gel purification kit.
- Digest Addgene plasmid #112688 with restriction enzyme AscI and XbaI24. In the resulting products, gel-purify the larger fragments, which is called the pre-base vector backbone, using gel purification kit.
- Assemble base vector with the pre-base vector backbone and the U6:3 fragment using the Gibson assembly method25. The base vector hereafter is named OA-1043.
- Synthesize target gene’s gRNA fragment using external gene synthesis service.
- Digest the base vector OA-1043 with restriction enzyme PstI and NotI24. Keep the entire digestion product, which iscalled digested OA-1043.
- Assemble gRNA expression vector with the digested OA-1043 and target gene gRNA fragment using the Gibson assembly method25.
NOTE: The Addgene ID of the resulting plasmid (OA-1052K) is #132422.
- Generating transgenic flies
- Inject OA-1052B vector into fly embryos using embryos from flies containing ØC31 integration site on the 3rd chromosome, BDSC fly stock number 9744, via external fly embryo injection service. Similarly, inject OA-1052K vector into fly embryos using embryos from flies containing ØC31 integration site on the 3rd chromosome, BDSC fly stock number 8622. Rear the injected embryos at 26 °C.
- Keep the dual-luciferase-expressing flies and the gRNA flies as homozygous lines; keep the Ubiq-CasRx lines as double-balanced heterozygous lines by outcrossing the single-balanced heterozygous Ubiq-CasRx line generated in section 1 to balancer lines carrying TM6 balancer chromosome with stubble (Stb) marker and retain only the double-balanced progenies with white-eyed, curly wings, and dsRed-fluorescent phenotypes simultaneously.
NOTE: The BDSC fly stock numbers are: #84127 (Ubiq-Fluc-Rluc), #84125 (gRNAFluc).
- Fly Genetics (Figure 1B and Figure 3A)
- Collect 8-10 virgin adult female flies from the dual-luciferase-expressing line. Collect 4-5 adult male flies from the balanced heterozygous Ubiq-CasRx/CyO; +/TM6, Stb line that show white-eyed, curly wings, and dsRed fluorescence simultaneously. Put the collected female and male flies, which are the parental flies, in a vial supplemented with dry yeast powder (hereafter called Step 1 Cross).
- Repeat the previous step 3 times for 3 replicates. For the control group, use the Ubiq-dCasRx/CyO; +/TM6, Stb line while keeping everything else the same.
- Rear the Step 1 Cross vials containing the parental flies at 26 °C for 48 hours. Then remove all parental flies from every vial. Then keep the vials at 26 °C for at least 14 days. During this time, collect 8-10 female virgins from the homozygous firefly luciferase-targeting gRNA line. Repeat this step 3 times for 3 replicates.
- Observe the Step 1 Cross vials every day to see if any new adult fly emerges from pupae. If so, anesthetize them with carbon dioxide, collect 5 male flies expressing both the Ubiq-CasRx (or Ubiq-dCasRx) and the dual-luciferase reporter from the progenies and put them into a new vial along with 10 virgin females from the firefly luciferase-targeting gRNA line (hereafter called Step 2 Cross). Repeat this step 3 times for 3 replicates.
- Collect another 5 one-day old male expressing both the Ubiq-CasRx (or Ubiq-dCasRx) and the dual-luciferase reporter from the Step 1 Cross vials and incubate them for 2 – 4 days at 26 °C. Then, transfer them into 1.5 mL centrifuge tube and store them at -80 °C. Repeat this step 3 times for three replicates.
- Rear the Step 2 Cross vials containing the parental flies at 26 °C for 48 hours. Then remove all parental flies from every vial. Then keep the vials at 26 °C for at least 20 days.
- Observe the Step 2 Cross vials every day to see if any new adult progenies emerged from pupae. If so, anesthetize them with carbon dioxide, score the anesthetized flies’ phenotypes and image them using color camera equipped with a fluorescent stereomicroscope. Count the numbers of progenies with different phenotypes. Use image processing software for image post-processing and compilation (Figure 3B – 3C).
NOTE: Mendelian genetics suggest that, if flies are all viable, 4 types of flies are expected among the progenies from Step 2 Cross, each accounting for 25% of the population (Figure 1B and Figure 3A).
- Luciferase assay (Figure 3D)
- Generate the triple transheterozygous flies as well as the control flies by repeating steps 2.4.1-2.4.5. Collect male flies at birth and age them until 3 days old.
- Transfer the 3-day old flies into 1.5 mL centrifuge tubes and lyse them using a pestle and the luciferase lysis buffer of commercially available luciferase assay kit.
- Use 5 µL of lysed tissue from each sample to measure both firefly and renilla luciferase activity using commercially available luciferase assay kit and luminometer.
3. Tissue-specific in vivo RNA Targeting Using a Three-Component CasRx System
- Generating UASt-CasRx and UASt-dCasRx Expression Vector
- PCR amplify the UASt promoter sequence using plasmid pJFRC81 and primers 1041.C9 and 1041.C11; then, PCR amplify CasRx fragment using plasmid OA-1050E (Addgene ID #132416) and primers 1050L.C1 and 1050E.C4; and then PCR amplify dCasRx fragments using plasmid OA-1050R (Addgene ID #132417) and primers 1050L.C1 and 1050E.C4 (Table 1)26. Gel-purify the amplified UASt promoter sequence, CasRx, and dCasRx fragments using gel purification kit.
- Digest base vector (Addgene plasmid #112686) with restriction enzymes NotI and PacI24. In the resulting products, gel-purify the larger fragment, which is called the base vector backbone, using gel purification kit.
- Assemble the UASt-CasRx vector with the base vector backbone, the UASt promoter sequence, and CasRx fragment using the Gibson assembly; then assemble the UASt-dCasRx vector with the base vector backbone, the UASt promoter sequence, and the dCasRx fragment using the Gibson assembly method25.
NOTE: The UASt-CasRx vector is Addgene plasmid #132418, and the UASt-dCasRx vector is Addgene plasmid #132419
- Generating transgenic flies
- Inject UASt-CasRx vector into fly embryos using fly embryo injection service and embryos from flies ØC31 integration site 8621 on their 2nd chromosomes; then inject UASt-dCasRx vector into fly embryos using fly embryo injection service and embryos from flies ØC31 integration site 8621 on their 2nd chromosomes. Rear the injected embryos at 26 °C.
- Keep the flies as double balanced heterozygous lines with CyO and Sb markers. NOTE: The IDs of the fly lines in BDSC are 84121 (UASt-CasRx) and 84120 (UASt-dCasRx).
- Fly genetics (Figure 1C)
- Order desired GAL4 lines from BDSC; Obtain relevant gRNA lines from step 3.2.2 (or from BDSC).
NOTE: The following 2 GAL4 flies from BDSC were used: GAL4-GMR (BDSC ID: #29967), GAL4-y (BDSC ID: #44373). The same 3 gRNA lines generated in the first section were used: gRNAw (BDSC ID: #84124), gRNAN (BDSC ID #84122), gRNAy (BDSC ID: #84123). - Collect 5-10 virgin adult female flies from the gRNA line. Collect 2-4 adult male flies from the double balanced heterozygous UASt-CasRx/CyO; +/TM6, Sb line that show white-eyed, curly wings, and dsRed fluorescence simultaneously. Put the collected female and male flies, which are the parental flies, in a regular food vial (hereafter called Step 1 Cross). Repeat this step 3 times for 3 replicates. For the control group, use the UASt-dCasRx/CyO; +/TM6, Sb line while keeping everything else the same.
- Rear the Step 1 Cross vials containing the parental flies at 26 °C for 48 hours. Then remove all parental flies from every vial. Then keep the vials at 26 °C for at least 14 days. During this time, collect 5-10 female virgins from the GAL4 line. Repeat this step 3 times for 3 replicates.
- Observe the Step 1 Cross vials every day to see if any new adult fly emerges from pupae. If so, anesthetize them with carbon dioxide, collect 2-4 male flies expressing both the UASt-CasRx (or UASt-dCasRx) and the gRNA vector from the progenies which simultaneously have dsRed-fluorescent and stubble phenotypes. Put the collected males from Step 1 Cross into a new vial along with 5-10 collected virgin female from the GAL4 line (hereafter called Step 2 Cross). Repeat this step 3 times for 3 replicates.
- Rear the Step 2 Cross vials containing the parental flies at 26 °C for 48 hours. Then remove all parental flies from every vial. Then keep the vials at 26 °C for at least 20 days.
- Observe the Step 2 Cross vials every day to see if any new adult fly emergs from pupae. If so, anesthetize them with carbon dioxide, score the anesthetized flies’ phenotypes and image them using color camera equipped with a fluorescent stereomicroscope. Count the numbers of progenies with different phenotypes. Use image processing software for image post-processing and compilation (Figure 4).
NOTE: Mendelian genetics suggest that, if flies are all viable, 4 types of flies are expected among the progenies from Step 2 Cross, each accounting for 25% of the population (Figure 1C).
- Order desired GAL4 lines from BDSC; Obtain relevant gRNA lines from step 3.2.2 (or from BDSC).
Results
Ubiquitous in vivo RNA Targeting Using a Two-Component CasRx System
The F1 transheterozygous flies expressing both the Ubiq-CasRx and the gRNA (targeting both endogenous and exogenous genes) constructs showed marked phenotypes compared to the control flies expressing the Ubiq-dCasRx and gRNA constructs (Figure 2 and Figure 4). Specifically, the transheterozygous CasRx flies have significantly lower levels of survival rate compared to the transheterozgyous dCasRx flies, indicating toxicity of the Ubiq-CasRx system (Figure 2A and Figure 4A). It is worth noting that both transheterozygous CasRx and dCasRx flies have less than 50% inheritance rate, which is the expected ratio based on Mendelian genetics. Of the three target genes, the Ubiq-CasRx/+; U6-gRNAN/+ flies and Ubiq-CasRx/+; U6-gRNAy/+ flies are non-viable (0% inheritance) and did not grow beyond the second instar larvae stage (Figure 2A-2B). The surviving Ubiq-CasRx/+; U6-gRNAw/+ flies, the inheritance of which was 12.9%, showed a distinct fully-penetrant white-eyed phenotype (Figure 2B). In addition to observable traits associated with CasRx, we were able to confirm significant reduction of target gene transcripts for 3 target genes: Notch, yellow, and GFP (Figure 2E-2G). Reduction of white gene transcripts was observed in Ubiq-CasRx/+, U6-gRNAw/+ flies, compared to the control Ubiq-dCasRx/+, U6-gRNAw/+ flies, though the reduction was not statistically significant (Figure 2E - 2F). Evidence of off-target activity induced by CasRx was found when comparing the differentially expressed transcripts between samples from CasRx-expressing flies and samples from dCasRx-expressing flies (Figure 2E, 2G). The number of non-target transcripts significantly differentially expressed are as follows: white, 253 (1.4% of total transcripts); Notch, 300 (1.7%); yellow, 41 (0.23%); GFP, 5,880 (33%) (Figure2G). Out of the total 17,779 different transcripts, 6 non-target transcripts were significantly differentially expressed in all 4 groups of samples. One of the 6 transcripts identified was Gadd45, a gene involved in apoptosis and cellular arrest in flies, raising the possibility that the enzymatic action of CasRx may either directly trigger cellular apoptosis or indirectly trigger misexpression of other genes, which in turn leads to apoptosis. Finally, it is worth noting that the Ubiq-CasRx and Ubiq-dCasRx flies were not established as homozygous stocks, presumably due to toxicity conferred by high ubiquitous expression. As a result, heterozygous Ubiq-CasRx/CyO and Ubiq-dCasRx/CyO flies were used for crossing with homozygous gRNA fly lines. In sum, the two-component Ubiq-CasRx system is able to achieve ubiquitous RNA targeting for both endogenous and exogenous targets resulting in observable phenotypes and transcript reduction. These results also showed that CasRx-mediated RNA targeting may introduce toxicity in vivo.
Ubiquitous in vivo exogenous RNA Targeting Using a Three-Component CasRx System
The results from the two-step cross showed that despite the exogenous nature of the target gene (i.e., Fluc), expressing all three transgenes in F2 triple transheterozygotes (Ubiq-CasRx/+; gRNAFluc/Ubiq-Fluc-Ubiq-Rluc) resulted in 100% lethality compared to control crosses involving Ubiq-dCasRx, where no lethality was observed in the F2 triple transheterozygotes (Ubiq-dCasRx/+; gRNAFluc/Ubiq-Fluc-Ubiq-Rluc) (Figure 3B-C). More specifically, only the combination of all three transgenes (Ubiq-CasRx/+; gRNAFluc/Ubiq-Fluc-Ubiq-Rluc) resulted in 100% lethality (Figure 3B and D), while (Ubiq-CasRx/+; gRNAFluc/TM6) and (Ubiq-CasRx/+; Ubiq-Fluc-Ubiq-Rluc/TM6) genotypes were viable and lacked phenotypes with their inheritance rates matching the expected Mendelian transmission rates, suggesting that the availability of the target sequence (i.e., firefly luciferase) in combination with Ubiq-CasRx/+ and the gRNAFluc is what resulted in the observed lethality phenotypes, presumably stemming from the collateral activity of Cas13 enzymes2,8. In addition, no distinguishable phenotypes or dramatic influence on inheritance in F1 transheterozygotes (Ubiq-CasRx/+; gRNAFluc/+ or Ubiq-CasRx/+; Ubiq-Fluc-Ubiq-Rluc/+) were observed compared to Ubiq-dCasRx controls (Ubiq-dCasRx/+; gRNAFluc/+ or Ubiq-dCasRx/+; Ubiq-Fluc-Ubiq-Rluc/+) (Figure 3B), indicating that a catalytically active enzyme is essential to obtain the lethality phenotypes observed. Furthermore, Fluc and Rluc expression levels in flies of all viable genotypes did not show significant reduction in Fluc expression in the Ubiq-dCasRx triple transheterozygotes (Ubiq-dCasRx/+; gRNAFluc/Ubiq-Fluc-Ubiq-Rluc) compared to dual luciferase reporter controls. This suggests that Fluc protein expression levels were not reduced by dCasRx targeting (Figure 3D). Taken together, the common lethality phenotype in the two different CasRx-mediated ubiquitous RNA targeting experiments indicate that when used on tissues ubiquitously, CasRx-mediated RNA targeting can be toxic to the organism.
Tissue-specific in vivo RNA Targeting Using a Three-Component CasRx System
The high level of toxicity observed in ubiquitous RNA targeting experiments prompted us to explore the tissue-specific RNA targeting using a three-component CasRx system design detailed in the methods section. Indeed, the level of toxicity observed was reduced when the overall CasRx expression was lowered using the UASt promoter compared to that of the Ubiq promoter, this is exemplified in three aspects: 1) the UASt-CasRx and UASt-dCasRx lines were maintained as homozygous lines, though based on the two-step cross scheme double balanced UASt-CasRx and UASt-dCasRx lines were used to perform the crosses, 2) all F2 generation dCasRx triple transheterozygous inheritance rates matched the expected 25% Mendelian inheritance rate, and 3) the F2 generation CasRx triple transheterozygous lethality phenotype was moderately reduced. In the white targeting experiment, of the 25% Mendelian inheritance rates expected in the F2 triple transheterozygotes, only 0.57% viable adult flies (UASt-CasRx/+; gRNAw/GMR-Gal4) were observed, all of which displayed severe eye specific pigmentation and morphology phenotypes (Figure 4A and 4B). For the white-targeting cross, the CasRx-expressing triple transheterozygous F2 inheritance rate was significantly lower than that of the dCasRx-expressing triple transheterozygous control group (27.6%) (Figure 4A). In the Notch targeting experiment, CasRx-expressing triple tranheterozygous carrying all three transgenes were 100% lethal, while the dCasRx control inheritance rate was 29.3% (Figure 4A). In the yellow targeting experiment, F2 triple transheterozygous CasRx-expressing, gRNAy, and y-GAL4 showed marginal chitin pigment reduction as small patches of yellow cuticle on the thorax and abdomen with an inheritance rate of 2.67%, much lower than that of the dCasRx control group (25.2%) (Figure4A). All dCasRx control triple transheterozygous flies did not present obvious phenotypes as the CasRx-expressing flies, indicating that catalytic activity of CasRx contributed to the phenotypes observed. The low inheritance rate in the CasRx triple transheterozygous group suggested that two sources of toxicity exist in CasRx RNA targeting: one is associated with high expression of CasRx, the toxicity of which was reduced by restrictive CasRx expression, the other is associated with the collateral activity. Taken together, these results showed that the CasRx system can achieve tissue-specific in vivo RNA targeting by leveraging the classical Gal4/UASt system and in the meantime reduce the toxicity. However, toxicity and occasional lethality phenotypes were still observed at a lower level of severity compared to that of the ubiquitous approaches, indicating that collateral cleavage activity is associated with toxicity.
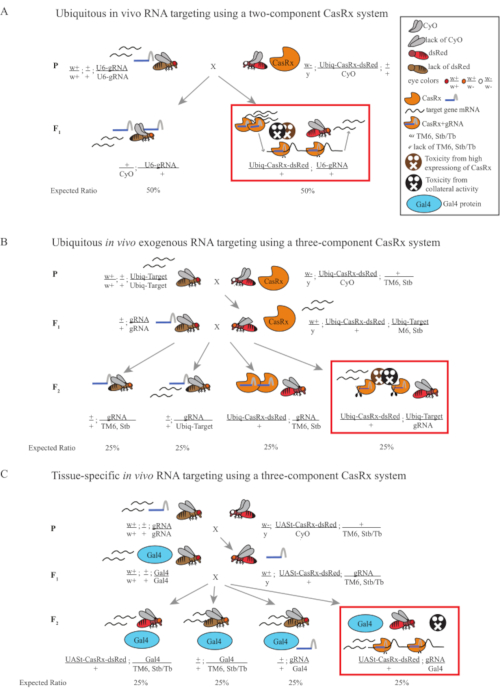
Figure 1: General overview of RNA targeting using a Cas13D system. (A) Schematics of the one-step genetic cross in the ubiquitous in vivo RNA targeting using a two-component CasRx system. (B) Schematics of a two-step genetic cross in the ubiquitous in vivo exogenous RNA targeting using the three-component CasRx system. (C) Schematics of a two-step genetic cross in the tissue-specific in vivo RNA targeting using a three-component CasRx system. Please click here to view a larger version of this figure.
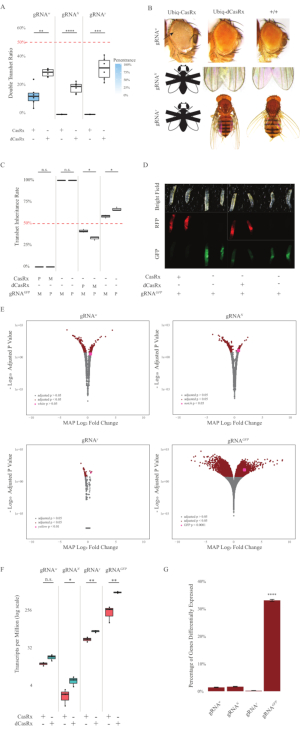
Figure 2: Ubiquitous in vivo RNA targeting using a two-component CasRx system (reprinted5). (A) Total inheritance percentages of transheterozygous flies inheriting Ubiq-CasRx (or Ubiq-dCasRx) and gRNAs. Blue shading in the box plot indicates phenotype penetrance. (B) Phenotypes of transheterozygous flies. Arrows indicate tissue necrosis in the eye. Black and white fly marked with ''X'' represents lethality. (C) Total inheritance percentages of transheterozygous flies of bidirectional crosses between Ubiq-CasRx (or Ubiq-dCasRx) and gRNAGFP-OpIE2-GFP flies. M, maternal inheritance of CasRx; P, paternal inheritance of CasRx. (D) F1 larvae progenies in the paternal cross. (E) Transcripts' maximum a posteriori estimates for the logarithmic fold change. DESeq2 pipeline was used. (F) Transcripts per million (TPM) targeted with CasRx or dCasRx. (G) CasRx-depentent differentially expressed transcript percentage of transcripts. Please click here to view a larger version of this figure.
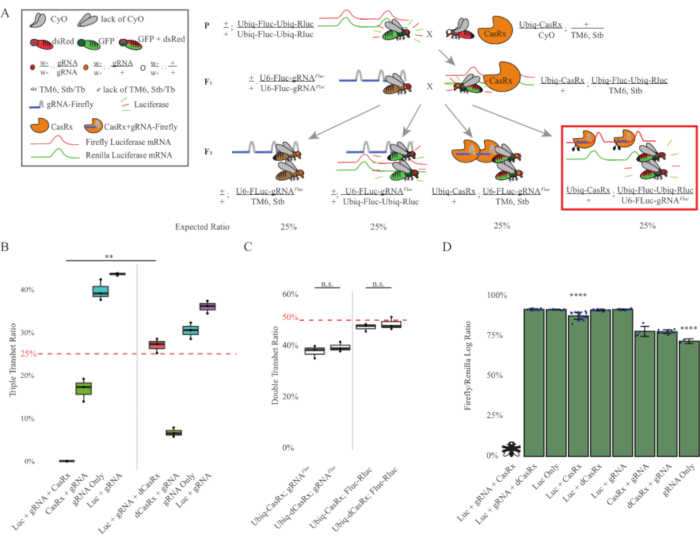
Figure 3: Ubiquitous in vivo exogenous RNA targeting using a three-component CasRx system. (A) Schematics of the two-step genetic cross. (B) Total inheritance percentages for all genotypes emerging in the F2 generation. Inheriting all three transgenes (Ubiq-CasRx, Ubiq-Fluc-Ubiq-Rluc, and gRNAFLuc) in F2 progeny resulted in 100% lethality and was significantly lower compared to the Ubiq-dCasRx triple transheterozygotes control group (p = 0.001, t-test). (C) Carrying Ubiq-CasRx/gRNAFluc alone or Ubiq-CasRx and Ubiq-Fluc-Ubiq-Rluc alone did not lead to severe lethality, and inheritance ratios between Ubiq-CasRx and Ubiq-dCasRx transheterozygotes were not significantly different (p = 0.41 and p = 0.51, respectively, t-test). (D) Luciferase ratios normalizing Fluc readings to Rluc readings. Triple transheterozygous flies expressing Ubiq-CasRx, Ubiq-Fluc-Ubiq-Rluc, gRNAFLuc were embryonic lethal, which was represented by a fly with an "X", and as a result luciferase expression was not measured. Fluc/Rluc ratio of Ubiq-CasRx/+, Ubiq-Fluc-Ubiq-Rluc/TM6, Stb transheterozygotes was significantly lower than that of the other Ubiq-Fluc-Ubiq-Rluc-expressing groups (p = 1.2e-06 or lower, t-test). The results from the gRNAFLuc-only group were significantly lower than that of all other groups (p = 1.2e-06 or lower, t-test). Please click here to view a larger version of this figure.
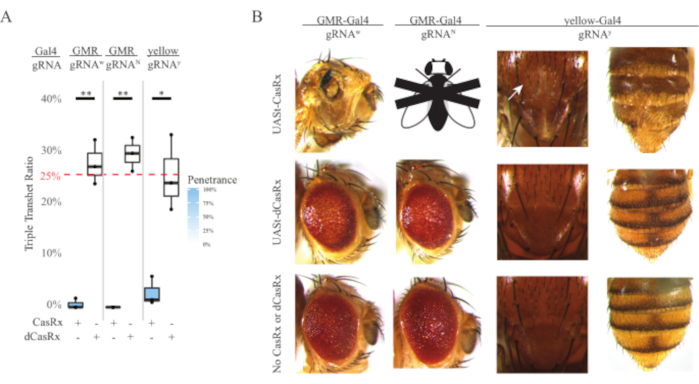
Figure 4: Tissue-specific in vivo RNA targeting using a three-component CasRx system (reprinted5). (A) Total inheritance percentage of triple transheterozygous flies carrying three transgenes (UASt-CasRx or UASt-dCasRx, gRNAs, and Gal4-driver. (B) Phenotypes of the triple transheterozygous flies. The white arrow indicates chitin pigment reduction in the thorax. Black and white fly marked with ''X'' represents lethality. Please click here to view a larger version of this figure.
| Construct | Description | Primer | Primer Sequence (5’ to 3’) | PCR Template |
| OA-1050E | CasRx | 1050E.C3 | TACTAATTTTCCAC ATCTCTATTTTGAC CCGCAGATTAATTA ATGAGCCCCAAGA AGAA | pNLS-RfxCas13d-NLS-HA (pCasRx) |
| 1050E.C4 | CAATTGATTTGTTA TTTTAAAAACGATT CATTCTAGCTAGCT TAAGCGTAATCTGG AACA | |||
| OA-1050R | dCasRx | 1050E.C3 | TACTAATTTTCCAC ATCTCTATTTTGAC CCGCAGATTAATTA ATGAGCCCCAAGA AGAA | pNLS-dRfxCas13d-NLS-HA (pdCasRx) |
| 1050E.C4 | CAATTGATTTGTTA TTTTAAAAACGATT CATTCTAGCTAGCT TAAGCGTAATCTG GAACA | |||
| OA-1050L | UASt promoter | 1041.C9 | GCGGGTTCTCGA CGGTCACGGCGG GCATGTCGACGC GGCCGCAACCAA CAACACTAGTAG | pJFRC81 |
| 1041.C11 | CTGGCCTCCACC TTTCTCTTCTTCT TGGGGCTCATGT TTAAACCCAATT CCCTATTCAGA | |||
| CasRx | 1050L.C1 | AATACAAGAAGA GAACTCTGAATA GGGAATTGGGT TTAAACATGAGC CCCAAGAAGAA | pCasRx | |
| 1050E.C4 | CAATTGATTTGT TATTTTAAAAAC GATTCATTCTA GCTAGCTTAAG CGTAATCTGGA ACA | |||
| OA-1050S | UASt promoter | 1041.C9 | GCGGGTTCTC GACGGTCACG GCGGGCATGT CGACGCGGCC GCAACCAACAA CACTAGTAG | pJFRC81 |
| 1041.C11 | CTGGCCTCCA CCTTTCTCTTC TTCTTGGGGCT CATGTTTAAAC CCAATTCCCTA TTCAGA | |||
| dCasRx | 1050L.C1 | AATACAAGAAG AGAACTCTGAAT AGGGAATTGGG TTTAAACATGAG CCCCAAGAAGAA | pdCasRx | |
| 1050E.C4 | CAATTGATTTGT TATTTTAAAAAC GATTCATTCTAG CTAGCTTAAGCG TAATCTGGAACA | |||
| OA-1043 | U6:3 promoter | 1043.C1 | GGGAATTGGGA ATTGGGCAATAT TTAAATGGCGGC GCGCCGAATTCT TTTTTGCTCACCT | Addgene plasmid #164586 |
| 1043.C23 | ACACTAGTGGAT CTCTAGAGGTAC CGTTGCGGCCG CAAAAAAGTTGT AATAGCCCCTCA AAACTGGACCTT CCACAACTGCAG CCGACGTTAAAT TGAAA | |||
| OA-1052B | Ubiq promoter | 1052B.C1 | GGGAATTGGGCA ATATTTAAATGGC GGCTGCAGCGC GCAGATCGCCGAT | Addgene plasmid #112686 |
| 1052B.C2 | TTTCTTTATGTTT TTGGCGTCTTCC ATCCTAGGTCTG CGGGTCAAAATA GAGATG | |||
| T2A-eGFP | 908A1 | ATAAAGGCCAAG AAGGGCGGAAA GATCGCCGTGG AGGGCAGAGGA AGTCTTCTAACAT GC | Addgene plasmid #112686 | |
| 908A2 | TTGTTATTTTAAAA ACGATTCATTCTA GGCGATCGCTTA CTTGTACAGCTC GTCCATGCC | |||
| Reversed Ubiq promoter | 908A3 | ACCGTGACCTAC ATCGTCGACACTA GTGGATCTCTAGA CGCGCAGATCGC CGATG | Addgene plasmid #112686 | |
| 908A4 | GGATCATAAACTT TCGAAGTCATGC GGCCGCTCTGCG GGTCAAAATAGAG ATGT |
Table 1: List of molecular construct and primers used in this study. This list includes all constructs (both the ID and description) and each construct's associated primers (both the ID and sequences (5' to 3')) and templates used.
Discussion
With three different application designs of the CasRx system, this work demonstrated in vivoprogrammable RNA targeting in flies. The different strategies cater to different project needs, such as endogenous versus exogenous gene targeting and ubiquitous versus tissue-specific RNA targeting. The effects of RNA targeting included target gene specific phenotypic changes, target RNA transcript reduction, and occasional lethality phenotypes associated with high expression of CasRx protein and collateral activity. Overall, these results showed that the CasRx system is capable of target RNA transcript reduction at the organismal level in a programmable and efficient manner.
One of the key factors in successful customizing of the CasRx system is the design of gRNAs. Specifically, the following advice shall be heeded: the target sequence is around 30 nucleotides in length, the length of poly-U stretches in the target sequence is 4 base pairs or less, the target sequence GC content is in the range of 30% - 70%, the target sequence is not predicted to form strong RNA hairpin structures, and the target sequence contains minimal predicted RNA secondary or tertiary structure5.
In addition to the gRNA designs, the fly genetics step in each protocol is also critical in a successful implementation. The presence or lack of the defined phenotypes passed down from the parents in the progenies are important for identifying and quantifying phenotypes induced by the CasRx system in the transheterozygous progenies. Also, setting up control crosses using the dCasRx flies in parallel are also helpful in the ruling out non-specific phenotypes in the transheterozygous progenies.
It is worth noting that these results revealed toxicity issue introduced by ubiquitously expressing CasRx and dCasRx protein in the fly, a limitation of CasRx system. Ubiquitous expression of CasRx or dCasRx under the Ubiq promotor alone, without gRNAs, came with nontrivial fitness costs, as neither Ubiq-CasRx nor Ubiq-dCasRx flies could be established as homozygous lines. On the contrary, UASt-CasRx and UASt-dCasRx flies can be established as healthy homozygous stocks, though due to the design of the cross scheme they were kept as double-balanced stocks, a fact that supports the existence of toxicity induced by ubiquitous CasRx protein expression. Another piece of supporting evidence is that in control experiments involving dCasRx, which is catalytically inactive, the percentages of flies carrying both dCasRx and gRNA constructs out of the total number of flies in the F1 generation were consistently lower than 50%, the ratio expected based on Mendelian genetics if no dCasRx-associated toxicity was present. This indicated that ubiquitously expressing dCasRx, along with gRNAs, induces toxicity in the fly, resulting less than expected inheritance ratio. The inheritance ratios of transheterozygous UASt-dCasRx, gRNA, GAL4 flies followed Mendelian genetics, which again suggests the toxicity induced specifically by ubiquitous expression of CasRx and dCasRx proteins. Toxicity in CRISPR/Cas system is not new. High amounts of Cas9 protein has been shown to be toxic in several organisms, including flies29,30,31,32. A recent study has developed a customized GAL4/UAS system that can tune the amount of Cas9 protein expressed in flies by adding an open-reading frame of varying length between the UAS sequence and the Cas9 sequence in the UAS-Cas9 construct33. Therefore, it is worth exploring ways to reduce CasRx-induced toxicity by tuning the CasRx protein expression level.
Other than the toxicity induced by ubiquitous expression of CasRx and dCasRx proteins, the results also showed lethality linked to CasRx system's non-specific collateral off-target effects, a feature of many CRISPR systems1, 2, 7, 34. In some of the CasRx and non-essential gene gRNA-expressing double or triple transheterozygous flies, for example when targeting Notch, the transheterozygous CasRx flies have significantly lower levels of survival rate compared to the transheterozgyous dCasRx flies. In the RNA-seq analysis of these CasRx and gRNA-expressing transheterozygous flies, both the reduction of target gene transcript levels and the reduction of non-target gene transcripts were observed. These collateral effects were CasRx-dependent and target-dependent, as they were only observed in transheterozygous flies expressing both the CasRx protein and the gRNA. It is worth pointing out that one of the target genes, white, showed only a limited, non-statistically significant reduction in transcripts when the white gene was targeted by CasRx, which was in contrast to the clear pigment reduction phenotype. It is hypothesized that this may be due to the fact that 1) the timing of RNA-seq sample collection was not well aligned with the timing when the white gene reach its peak expression during early development, and 2) the localized expression of the white gene in the eyes makes it challenging to collect the relevant tissues during early development phase when only whole-body sample collection is feasible. To reduce collateral activity in the CasRx system, future studies are called for to fully understand the mechanisms underlying the off-target phenomenon system at the organismal level.
Interestingly, a recent study35 describing RNA-targeting Cas13 tools in flies appeared to ameliorate the general toxicity associated with CasRx expression, for several possible reasons. Firstly, the authors recoded the Cas13 transgenes to optimize expression in Drosophila and utilized a more weakly-expressing promoter (actin 5C) as compared to the ubiquitin promoter used in the present study, likely leading to lower levels of Cas13 expression and thus less toxicity. Indeed, this is supported by the observations that UASt-driven CasRx and dCasRx expression was not, by itself, toxic, as this study (and the authors in 35) did not observe any obvious lethality in UASt-CasRx flies. Furthermore, these authors encoded their gRNAs differently compared to this study, which may have affected their expression and reduced the toxicity of the system in transheterozygous Cas13/gRNA flies. For example, in their study two gRNAs were expressed using the U6:3 promoter and flanked by tRNAs to enable gRNA processing upon tRNA maturation without requiring CasRx35. Conversely, in this study, the gRNAs were encoded as arrays targeting up to 4 locations per gene and mimicking the endogenous Cas13 array structure found in bacteria, which requires the Cas13 enzyme to process each gRNA. These different approaches may have led to differences in gRNA expression levels and other factors that may have inherent effects on the toxicity of the whole system. Finally, Huynh et al. targeted different genes than those targeted in the present study, which result in differences in target-Cas/gRNA interaction and collateral activity and may have effects on the observed levels of lethality. These differences in observed toxicity warrant further investigation to identify ways that the overall systems can be improved.
Overall, this study is the first demonstration of a functional genetically encoded programmable RNA-targeting Cas system in D. melanogaster, albeit further optimization of the CasRx system (in line with what is reported35) will be required to further reduce the off-target-associated lethality and increase the efficacy of CasRx on-target cleavage. RNA-targeting with Cas enzymes is a rapidly evolving field with many potential applications ranging from insect vector control to therapeutic usages1,2,3,4,5,6,7, and this protocol offer a starter package for anyone interested in designing their first CasRx system in flies, while being compatible with customization and further optimization of the system. The examples presented here demonstrate a range of results one may encounter during implementation of this system in vivo and may serve as benchmarks for other users in evaluating the performance of CasRx system in their applications.
Disclosures
O.S.A is a founder of Agragene, Inc., has an equity interest, and serves on the company's Scientific Advisory Board. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. All other authors declare no competing interests.
Acknowledgements
This work was supported in part by funding from a DARPA Safe Genes Program Grant (HR0011-17-2-0047), and NIH awards (R21RAI149161A, DP2AI152071) awarded to O.S.A.
Materials
| Name | Company | Catalog Number | Comments |
| 100% Grape Juice | Welch Foods Inc. | N/A | |
| Active Dry Yeast (908g) | Red Star Yeast Company, LLC | N/A | |
| DMC4500 color camera | Leica Microsystems | DMC4500 | |
| Dual-Luciferase Reporter Assay System | Promega | E1910 | |
| GAL4-GMR flies | Bloomington Drosophila Stock Center | 29967 | |
| GAL4-y flies | Bloomington Drosophila Stock Center | 44373 | |
| Glomax 20/20 Luminometer | Promega | E5331 | |
| Illumina HiSeq2500 Sequencer | Illumina, Inc. | HiSeq2500 | |
| M165FC fluorescent stereomicroscope | Leica Microsystems | M165FC | |
| Nanodrop OneC UV-vis spectrophotometer | ThermoFisher | NDONEC-W | |
| NEBNext Ultra II RNA Library Prep Kit | New England Biolabs, Inc. | E7770 | |
| plasmid # 112686 | Addgene | 112686 | |
| plasmid # 112688 | Addgene | 112688 | |
| plasmid # 132416 | Addgene | 132416 | |
| plasmid # 132417 | Addgene | 132417 | |
| plasmid # 132419 | Addgene | 132419 | |
| plasmid # 132420 | Addgene | 132420 | |
| plasmid # 132421 | Addgene | 132421 | |
| plasmid # 132422 | Addgene | 132422 | |
| plasmid # 132425 | Addgene | 132425 | |
| plasmid # 132426 | Addgene | 132426 | |
| plasmid # 133304 | Addgene | 133304 | |
| plasmid # 164586 | Addgene | 164586 | |
| plasmid #132418 | Addgene | 132418 | |
| plasmid pJFRC81 | Addgene | 36432 | |
| Qiagen RNeasy Mini Kit | Qiagen | 74104 | |
| Restriction endonucleases AscI | New England Biolabs Inc. | R0558L | |
| Restriction endonucleases NotI | New England Biolabs Inc. | R0189L | |
| Restriction endonucleases PacI | New England Biolabs Inc. | R0547L | |
| Restriction endonucleases PstI | New England Biolabs Inc. | R0140L | |
| Restriction endonucleases SwaI | New England Biolabs Inc. | R0604L | |
| Restriction endonucleases XbaI | New England Biolabs Inc. | R0145L | |
| RNA 6000 Pico Kit for Bioanalyzer | Agilent Technologies | 5067-1513 | |
| Turbo DNase | Invitrogen | AM2238 | |
| U6-3:4-gRNA-Fluc flies | Bloomington Drosophila Stock Center | 84125 | |
| U6-3:4-gRNA-GFP; OpIE2-GFP flies | Bloomington Drosophila Stock Center | 84986 | |
| U6-3:4-gRNA-N flies | Bloomington Drosophila Stock Center | 84122 | |
| U6-3:4-gRNA-w flies | Bloomington Drosophila Stock Center | 84124 | |
| U6-3:4-gRNA-y flies | Bloomington Drosophila Stock Center | 84123 | |
| UASt-CasRx flies | Bloomington Drosophila Stock Center | 84121 | |
| UASt-dCasRx flies | Bloomington Drosophila Stock Center | 84120 | |
| Ubiq-CasRx flies | Bloomington Drosophila Stock Center | 84118 | |
| Ubiq-dCasRx flies | Bloomington Drosophila Stock Center | 84119 | |
| Ubiq-Firefly-T2A-eGFP-Ubiq-Renilla flies | Bloomington Drosophila Stock Center | 84127 | |
| Zymoclean Gel DNA Recovery Kits | Zymo Research Corporation | D4007 |
References
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat Communications. 9, 1911 (2018).
- Abudayyeh, O., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 353 (6299), 5573 (2016).
- East-Seletsky, A., O'Connell, M., Burstein, D., Knott, G., Doudna, J. RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Molecular Cell. 66 (3), 373-383 (2017).
- Konermann, S., Lotfy, P., Brideau, N., Oki, J., Shokhirev, M., Hsu, P. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 173 (3), 665-676 (2018).
- Buchman, A., Brogan, D., Sun, R., Yang, T., Hsu, P., Akbari, O. Programmable RNA Targeting Using CasRx in Flies. The CRISPR Journal. 3 (3), 164-176 (2020).
- Kushawah, G., et al. CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos. Developmental Cell. 54 (6), 805-817 (2020).
- Abudayyeh, O., et al. RNA targeting with CRISPR-Cas13. Nature. 550, 280-284 (2017).
- Perrimon, N., Ni, J., Perkins, L. In vivo RNAi: today and tomorrow. Cold Spring Harbor Perspectives in Biology. 2, 003640 (2010).
- Dietzl, G., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448, 151-156 (2007).
- Ni, J., et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 182 (4), 1089-1100 (2009).
- Ni, J., et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nature Methods. 8, 405-407 (2011).
- Ni, J., et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nature Methods. 5, 49-51 (2008).
- Heigwer, F., Port, F., Boutros, M. RNA Interference (RNAi) Screening in Drosophila. Genetics. 208 (3), 853-874 (2018).
- Kulkarni, M., et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nature Methods. 3, 833-838 (2006).
- Ma, Y., Creanga, A., Lum, L., Beachy, P. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 443, 359-363 (2006).
- Perrimon, N., Mathey-Prevot, B. Matter arising: off-targets and genome-scale RNAi screens in Drosophila. Fly. 1 (1), 1-5 (2007).
- Markstein, M., Pitsouli, C., Villalta, C., Celniker, S., Perrimon, N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nature Genetics. 40, 476-483 (2008).
- Champer, J., Buchman, A., Akbari, O. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nature Review Genetics. 17 (3), 146-159 (2016).
- Buchman, A., et al. Broad dengue neutralization in mosquitoes expressing an engineered antibody. PLoS Pathogens. 16 (4), 1008103 (2020).
- Mathur, G., Sanchez-Vargas, I., Alvarez, D., Olson, K., Marinotti, O., James, A. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Molecular Biology. 19 (6), 753-763 (2011).
- Franz, A., et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America. 103 (11), 4198-4203 (2006).
- Yen, P., James, A., Li, J., Chen, C., Failloux, A. Synthetic miRNAs induce dual arboviral-resistance phenotypes in the vector mosquito Aedes aegypti. Communications Biology. 1, 11 (2018).
- Buchman, A., et al. Engineered resistance to Zika virus in transgenic Aedes aegypti expressing a polycistronic cluster of synthetic small RNAs. Proceedings of the National Academy of Sciences of the United States of America. 116 (9), 3656-3661 (2019).
- . Addgene Available from: https://www.addgene.org/protocols/restriction-digest/ (2020)
- Gibson, D., Young, L., Chuang, R., Venter, J., Hutchison, C., Smith, H. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 6 (5), 343-345 (2009).
- . Addgene Available from: https://www.addgene.org/protocols/pcr/ (2020)
- . Indiana University Bloomington Available from: https://bdsc.indiana.edu/information/recipes/bloomfood.htmL (2020)
- Dobin, A., et al. STAR:ultrafast universal RNA-seq aligner. Bioinformatics. 29 (1), 15-21 (2013).
- Jiang, W., Brueggeman, A., Horken, K., Plucinak, T., Weeks, D. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryotic Cell. 13 (11), 1465-1469 (2014).
- Poe, A., et al. Robust CRISPR/Cas9-Mediated Tissue-Specific mutagenesis reveals gene redundancy and perdurance in Drosophila. Genetics. 211 (2), 459-472 (2019).
- Port, F., Chen, H., Lee, T., Bullock, S. Optimized CRISPR/Cas tools for efficient ger mLine and somatic genome engineering in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 111 (29), 2967-2976 (2014).
- Yang, S., Li, S., Li, X. Shortening the Half-Life of Cas9 maintains its gene editing ability and reduces neuronal toxicity. Cell Reports. 25 (10), 2653-2659 (2018).
- Port, F., et al. A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. eLife. 9, 53865 (2020).
- Zhang, X., Tee, L., Wang, X., Huang, Q., Yang, S. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy - Nucleic Acids. 4 (17), 264 (2015).
- Huynh, N., Depner, N., Larson, R., King-Jones, K. A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biology. 21, 279 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved