A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
DNA Tension Probes to Map the Transient Piconewton Receptor Forces by Immune Cells
In This Article
Summary
This paper describes a detailed protocol for using DNA-based tension probes to image the receptor forces applied by immune cells. This approach can map receptor forces >4.7pN in real-time and can integrate forces over time.
Abstract
Mechanical forces transmitted at the junction between two neighboring cells and at the junction between cells and the extracellular matrix are critical for regulating many processes ranging from development to immunology. Therefore, developing the tools to study these forces at the molecular scale is critical. Our group developed a suite of molecular tension sensors to quantify and visualize the forces generated by cells and transmitted to specific ligands. The most sensitive class of molecular tension sensors are comprised of nucleic acid stem-loop hairpins. These sensors use fluorophore-quencher pairs to report on the mechanical extension and unfolding of DNA hairpins under force. One challenge with DNA hairpin tension sensors is that they are reversible with rapid hairpin refolding upon termination of the tension and thus transient forces are difficult to record. In this article, we describe the protocols for preparing DNA tension sensors that can be "locked" and prevented from refolding to enable "storing" of mechanical information. This allows for the recording of highly transient piconewton forces, which can be subsequently "erased" by the addition of complementary nucleic acids that remove the lock. This ability to toggle between real-time tension mapping and mechanical information storing reveals weak, short-lived, and less abundant forces, that are commonly employed by T cells as part of their immune functions.
Introduction
Immune cells defend against pathogens and cancer cells by continuously crawling and scanning the surfaces of target cells for antigens, studding their surface1,2. Antigen recognition is initiated upon binding between the T cell receptor (TCR) and the peptide-major histocompatibility complex MHC (pMHC) complex expressed on the surface of target cells. Because TCR-pMHC recognition occurs at the junction between two mobile cells, it has long been suspected of experiencing mechanical forces. Moreover, this led to the mechanosensor model of TCR activation, which suggests that TCR forces contribute to its function3,4. To understand when, where, and how mechanical forces contribute to T cell function, it is imperative to develop tools to visualize the molecular forces transmitted by T cells. Traditionally, methods such as traction force microscopy (TFM) and micropillar arrays are used to investigate cellular forces5,6. However, the force sensitivity of TFM and micropillar arrays is at the nanonewton (nN) scale and thus is often insufficient to study the molecular piconewton (pN) forces transmitted by cell receptors7. To improve the force and spatial resolution for detection, our lab pioneered the development of molecular tension probes, which were initially synthesized using polyethylene glycol (PEG) polymers7. Molecular tension probes are comprised of an extendible molecular "spring" (PEG, protein, DNA) flanked by a fluorophore and quencher and are anchored on a surface. Forces applied to the terminus of the probe lead to its extension, separating the fluorophore and quencher, and thus generating a strong fluorescence signal (Figure 1A)8,9,10.
Over the past decade we have developed a library of different classes of molecular tension probes with spring elements made from nucleic acids11, proteins10, and polymers8. Among these, the DNA-based tension probes provide the highest signal to noise ratio and the greatest force sensitivity, which is easily tuned from a few pN up to ~20 pN11. We have used these real-time DNA tension probes to study the molecular forces generated by many diverse cell types, including fibroblasts, cancer cells, platelets, and immune cells11,12,13. This manuscript will describe protocols to synthesize and assemble DNA tension probes on a surface to map molecular receptor forces with pN force resolution using a conventional fluorescence microscope. While the current procedure includes chemical modifications to the nucleic acid to introduce the fluorescent reporter (Figure 1B), it is important to note that many of the modification and purification steps can be outsourced to custom DNA synthesis companies. Therefore, DNA tension probes technology is facile, and accessible to the broader cell biology and mechanobiology communities.
Briefly, to assemble DNA tension sensors, a DNA hairpin is hybridized to a fluorescent ligand strand on one arm and a quencher anchor strand on the other arm and then immobilized on a glass substrate (Figure 1C, real-time tension). In the absence of mechanical force, the hairpin is closed, and thus the fluorescence is quenched. However, when the applied mechanical force is greater than the F1/2 (the force at equilibrium that leads to a 50% probability of unfolding), the hairpin mechanically melts, and a fluorescent signal is generated.
Building on the real-time DNA tension sensor, we also describe protocols to map accumulated forces, which is particularly useful for studying interactions between receptors on immune cells and their natural ligand. This is because immune receptors often display short-lived bonds3,14. Accumulated forces are imaged using a "locking" strand that preferentially binds to open DNA hairpins and allows for the storage of fluorescence signals associated with mechanical pulling events (Figure 1C, locked tension). The locking strand is designed to bind a cryptic binding site that is exposed upon mechanically induced melting of the hairpin and lock the hairpin in the open state by blocking hairpin refolding, thus storing the tension signal, and generating an accumulated tension map. Moreover, the locking strand is designed with an eight-nucleotide toehold, which enables a toehold-mediated strand displacement reaction with its full complement, the "unlocking" strand. With the addition of the unlocking strand, the bound locking strand is stripped off the hairpin construct, erasing the stored tension signal and resetting the hairpin back to the real-time state.
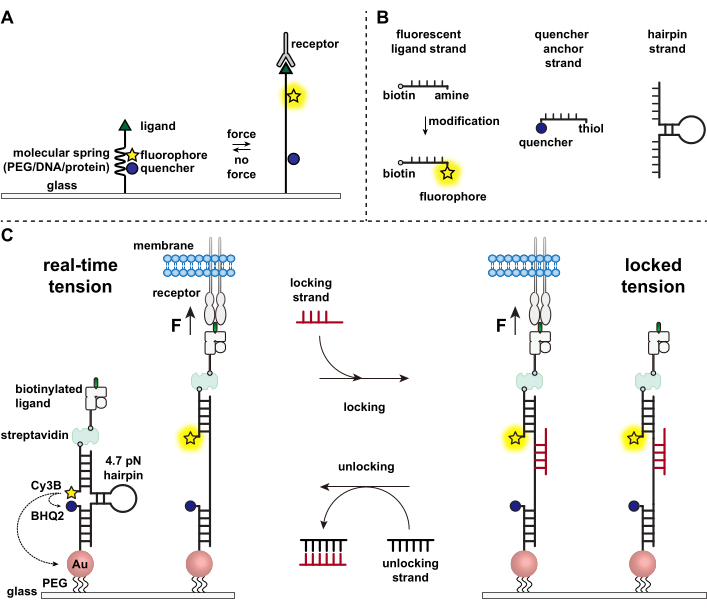
Figure 1: Scheme of the state-of-art molecular tension probes. (A) General design of real-time molecular tension probe, (B) Strands for the DNA-based tension probe construct, and (C) engineered DNA-based tension probes and their toggling between real-time state and locked state. Please click here to view a larger version of this figure.
The main protocol consists of four major sections - oligonucleotide preparation, surface preparation, imaging, and data analysis. This protocol has been successfully demonstrated by our lab and others in naïve and activated OT-1 CD8+ T cells, OT-II CD4+ cells, as well as hybridomas, and can be applied to interrogate different immune cell receptors including T cell receptor, programmed cell death receptor (PD1), and lymphocyte function-associated antigen 1 (LFA-1) forces. OT-1 CD8+ naïve T cells are used as an example cell line in this paper.
Protocol
The OT-1 transgenic mice are housed at the Division of Animal Resources Facility at Emory University. All the experiments were approved and performed under the Institutional Animal Care and Use Committee (IACUC) protocol.
1. Oligonucleotide preparation
- Dissolve the ligand strand DNA in water (18.2 MΩ resistivity, used throughout the whole protocol). Vortex and spin down the solution with a tabletop centrifuge. Tune the volume of water such that the final concentration is 1 mM. Validate the concentration by using a nanodrop spectrophotometer to measure the absorbance at 260 nm and determine the final concentration based on the extinction coefficient of the oligonucleotide.
NOTE: The ligand strand has a modification at each terminus, 5' amine and 3' biotin, to conjugate with the fluorophore and to present the biotinylated ligand. The amine group in the ligand strand needs to be conjugated with a fluorophore. Cy3B dye is used for this conjugation due to its high brightness and photostability, but it is not generally offered commercially and requires in-house conjugation. Accordingly, the following section describes the conjugation between amines and NHS ester dyes. For end users that do not have access to facilities or resources for nucleic acid modification, modified nucleic acids can instead be purchased from custom DNA synthesis vendors that offer bright and photostable dyes, such as the Alexa and Atto family of dyes. - Prepare 10x PBS and 1 M NaHCO3 solutions. Mix 10 µL of the 1 mM amine ligand strand solution (10 nmol) with 10 µL of 10x PBS, 10 µL of 1 M NaHCO3, and 60 µL of H2O. Dissolve 50 µg of Cy3B NHS ester in 10 µL of DMSO immediately before use and add to the mixture for a total reaction volume of 100 µL. Add Cy3B NHS ester last. Allow to react at room temperature for 1 h or 4 °C overnight.
- Prepare the Atto647N locking strand by conjugating the amine locking strand with Atto647N NHS ester. Prepare 10x PBS and 1 M NaHCO3 solution. Mix 10 µL of the 1 mM amine locking strand solution (10 nmol) with 10 µL of 10x PBS, 10 µL of 1 M NaHCO3, and 60 µL of H2O. Dissolve 50 µg of Atto647N NHS ester in 10 µL of DMSO immediately before use and add to the mixture for a total reaction volume of 100 µL. Add Atto647N NHS ester last. Allow to react at room temperature for 1 h or 4 °C overnight.
- After the reactions, remove by-products, excess dye, and salts by P2 desalting gel filtration. Dilute the reaction mixture with H2O to a total volume of 300 µL, which is appropriate for the subsequent HPLC purification step. Add 650 µL of hydrated P2 gel to a centrifugal device and spin down at 18,000 x g for 1 min. Remove the liquid at the bottom of the device, add the reaction mixture to the column containing P2 gel, spin down at 18,000 x g for 1 min and collect the reaction mixture at the bottom of the device.
NOTE: P2 gel should be hydrated at least 4 h before use with H2O. - Purify the desalted reaction mixture with HPLC using a C18 column designated for oligonucleotide purification, with solvent A: 0.1 M TEAA in H2O and B: ACN as the mobile phase for a linear gradient elution 10-100% B over 50 min at a flow rate of 0.5 mL/min. Inject the desalted reaction mixture in reverse-phase HPLC with a 500 µL injection loop for purification. Collect the product that has an absorbance peak for the DNA (260 nm) and an absorbance peak for the fluorophore (560 nm for Cy3B and 647 nm for Atto647N) and dry them in a vacuum centrifugal concentrator overnight (see Figure 2A).
- Reconstitute the dried oligo-dye product in 100 µL water. Determine the concentration of the Cy3B ligand strand and Atto647N locking strand with the nanodrop spectrophotometer. Ensure that the dye labeling ratio is close to 1:1. Correct for the 260 nm absorbance of the dye if needed when determining the oligonucleotide concentration.
- Validate the purified product with MALDI-TOF-MS using 3-HPA as the substrate in 50% ACN/H2O with 0.1% TFA and 5 mg/mL ammonium citrate using 0.5 µL of the product at 1-5 µM for MALDI-TOF-MS sample preparation. An example mass spectrum can be found in Figure 2B.
- Dissolve the hairpin strand and the quencher anchor strand in water and make sure the concentration of the stock solutions is between 50 and 100 µM.
NOTE: The hairpin strand is unmodified and can be directly custom synthesized from a vendor. The anchor strand has a thiol anchoring group and a quencher BHQ2 and can be directly custom synthesized from a vendor. - Aliquot all of the oligonucleotides. For short-term use and storage, store these oligonucleotides at 4 °C. For long-term storage, freeze and keep them at -20 °C. At this point, all of the oligonucleotides are ready for the DNA tension probe assembly.
NOTE: Repeated freeze-thaw cycles are not problematic for oligonucleotides.
2. Surface preparation
NOTE: The preparation of DNA hairpin tension probe substrates takes two days. The DNA hairpin tension probe will be functionalized onto glass coverslips.
- Day 1
- Place the 25 mm coverslips on a polytetrafluoroethylene rack in a 50 mL beaker. Each rack can hold up to 8 coverslips. Rinse the coverslips by submerging in water three times.
- Add 40 mL of a 1:1 ratio (v:v) solution of ethanol mixed with water to the beaker containing the rack and coverslips, and seal the beaker using a paraffin film.
- Sonicate the beaker for 15 min in an ultrasonics cleaner (operating frequency 35 KHz) to clean the coverslips. After sonication, discard the liquid and rinse the beaker with the rack and coverslips in it with water at least 6 times to remove any remaining organic solvent.
- Prepare fresh Piranha solution by mixing sulfuric acid and hydrogen peroxide in the ratio of 3:1. To make 40 mL of Piranha solution, add 30 mL of sulfuric acid to a clean 50 mL beaker first and then slowly add 10 mL of H2O2. The Piranha solution will rapidly heat and bubble upon addition of the H2O2. Gently mix the piranha using the end of a glass pipette.
- Next, transfer the rack that holds the coverslips to the beaker containing gently mixed Piranha solution for etching (Figure 3A). Allow the Piranha solution to hydroxylate and clean the coverslips for 30 min at room temperature. After Piranha etching, transfer the rack using steel or polytetrafluoroethylene tweezers to a clean 50 mL beaker with water and rinse again with water at least 6 times.
CAUTION: Large amounts of organic substances could react vigorously with Piranha solution and may cause explosion. Be careful and always work with Piranha solution in a fume hood. Make sure to wear a labcoat, gloves, and safety goggles. Never store fresh Piranha solution in a sealed container.
NOTE: The hydrogen peroxide to sulfuric acid ratio should be kept under 1:2 (v:v) and should never exceed 1:1. When submerging the rack with coverslips in Piranha solution, place them in the solution slowly and carefully. Do not discard the solution immediately after etching, as it is still active and hot. Leave it in the beaker overnight before pouring it in the acid waste container. - Immerse the rack holding the coverslips in a 50 mL beaker with 40 mL ethanol to remove water. Discard the ethanol and repeat 3 times to ensure that the water has been removed.
- Then immerse the rack in 3% aminopropyl triethoxy silane (APTES) (v/v) in 40 mL of ethanol to react with the -OH on the coverslips for 1 h at room temperature (Figure 3B).
NOTE: Ethanol can be replaced by acetone. - Rinse surfaces 6 times by submerging them into 40 mL ethanol, then dry in oven at 80 °C for 20 min. After cooling, store the dried amine-modified coverslips at -20 °C for future use (up to 6 months).
- Cover the bottom inner side of 10 cm diameter plastic Petri dishes with paraffin film. The paraffin film prevents the coverslips from sliding inside the Petri dish and helps in keeping the solution for the next steps of functionalization stay on the coverslips. Place the cooled-down amine-modified coverslips in the Petri dishes. The side to be functionalize should be facing up.
- To modify the amine groups on the coverslips, add 300 µL of 0.5% w/v lipoic acid PEG NHS (LA-PEG-SC) and 2.5% w/v mPEG NHS (mPEG-SC) in 0.1 M NaHCO3 onto each coverslip and incubate for 1 h at room temperature (Figure 3C). For each 25 mm coverslip, weigh 1.5 mg of LA-PEG-SC and 7.5 mg of mPEG-SC. Dissolve the NHS reagents immediately before adding to the surfaces, as they have a short half-life (~10 min) in aqueous solution at room temperature. After the reaction, rinse surfaces 3 times with water.
NOTE: The NHS reaction can be performed at 4 °C overnight. NHS reagents have longer half-life before hydrolysis at 4 °C, which is around 4-6 h. This will result in a three-day surface prep procedure. - Add 100 µL of 0.1 M NaHCO3 containing 1 mg/mL of sulfo-NHS acetate to a set of "sandwich" coverslips (two coverslips facing towards each other with reaction buffer in between). Allow passivation to occur for at least 30 min. To save reagent, this step could be done with 50 µL of 1 mg/mL sulfo-NHS acetate. Rinse with water three times after passivation.
- Add 0.5 mL of gold nanoparticles (AuNP, 8.8 nm, tannic acid, 0.05 mg/mL) to each coverslip and incubate for 30 min at room temperature (Figure 3D). To save the reagent, this step can be done by sandwiching two coverslips as well. Make sure no salts are present in the system from previous steps to avoid aggregation of gold nanoparticles. Do not leave the coverslips to dry after this step.
- Meanwhile, pre-hybridize 4.7 pN hairpin, Cy3B ligand strand, and BHQ2 anchor strand that form the DNA tension probes construct at a ratio of 1.1:1:1 in 1 M NaCl at 300 nM in a PCR tube. Anneal the strands by heating the solution up to 95 °C for 5 min, then gradually cool down by decreasing the temperature to 20 °C over 30 min in a thermal cycler.
- Rinse the coverslips with water three times after 30 min of incubation with gold nanoparticles. Add additional BHQ2 anchor strand (from 100 µM stock) to the annealed DNA solution to make the ratio between BHQ2 anchor strand and Cy3B ligand strand 10:1. At this point, the DNA solution should contain 300 nM of tension probe construct and 2.7 µM BHQ2 strand. Add 100 µL per two coverslips to make the "sandwich" (Figure 3E).
- Carefully place a wet lab tissue ball in the Petri dish (away from coverslips) and seal the dish with paraffin film to prevent the solution from drying up. Cover the dish with foil and incubate at 4 °C overnight.
- Day 2
- Wash off the excess probes from the coverslips with 1x PBS. Check for DNA tension probe surface quality under an epifluorescent microscope.
- Prepare 40 µg/mL of streptavidin in 1x PBS and incubate on coverslips for 30 min at room temperature (Figure 3F). Usually, 100 µL is sufficient for a 25 mm coverslip. Rinse with PBS 3 times after incubation to wash away the excess amount of streptavidin.
- Prepare 40 µg/mL biotinylated antibody/ligand in 1x PBS. Add 50-100 µL per sandwich and incubate for 30 min at room temperature (Figure 3G). Rinse with PBS three times after incubation to wash away the excess amount of biotinylated antibody/ligand.
- Assemble the clean imaging chambers with surfaces carefully. Surfaces can be easily cracked when tightening the chambers. Add 0.5-1 mL of Hank's balanced salt solution (HBSS) to the imaging chambers and keep them ready for imaging with cells (Figure 3H).
3. Imaging cell receptor forces
- Prepare immune cells of interest in HBSS at 1-2 x 106 cells/mL.
NOTE: OT-1 CD8+ naïve cells are used as an example in this paper. Purify OT-1 CD8+ naïve T cells a from the spleens of sacrificed mice using the MACS mouse CD8+ T cell isolation kit with a MACS separator following manufacturer's instruction. Isolate and enrich the CD8+ T cells by removing any non CD8+ T cells that magnetic depleting antibody cocktail bound to. Resuspend purified OT-1 CD8+ naïve T cells in HBSS at 2 x 106 cells/mL and keep on ice prior to use. - Check the quality of DNA hairpin tension probe surface under a fluorescence microscope (100x objective) for quality control before adding ligands or plating cells. Image and quantify the average background intensity in Cy3B channel of a DNA hairpin tension probe surface from at least 5 different positions and 3 replicates. Keep the imaging acquisition conditions consistent so that this value can be used as a reliable marker of surface quality and probe density (Figure 4C).
NOTE: Quantify the number of DNA strands per gold nanoparticle and the number of gold nanoparticles per µm2 the first few times of surface preparation according to literature12, which can be used as another reliable marker of surface quality. - Plate ~4 x 104 - 10 x 104 cells onto each DNA tension probe functionalized coverslip and allow them to attach and spread for ~15 min at room temperature.
- As cells are plated onto the DNA hairpin tension probes and start to spread, image the fluorescence signals that are generated in the Cy3B channel with the 100x objective (Figure 3I).
- After cells start to produce real-time tension signal on the DNA hairpin tension probe surface in the Cy3B channel, acquire images in both Cy3B and Atto647N channels (TIRF microscopy gives better signal-to-noise ratio than epifluorescence). Subsequently, add Atto647N strand to the imaging chambers at a final concentration of 200 nM for mechanically selective hybridization.
- After 10 min of incubation, quickly and gently remove the buffer containing the fluorescent Atto647N locking strand and replace with fresh Hank's balanced salts. Image in both Cy3B and Atto647N channels again and determine the Pearson's correlation coefficient with Fiji software15.
- At the timepoint of interest for the investigation, introduce non-fluorescent locking strand to the cells in the imaging chamber to store the tension signal. Prepare locking strand stock (100 µM) and add to the cells at a final concentration of 1 µM. Gently pipette to mix. The duration of locking can vary but 10 min is the recommended time.
- Acquire time-lapse movies or end-point images in epifluorescence for both qualitative tension mapping and quantitative analysis as needed (Figure 3J and Figure 5).
NOTE: If tension measurement at multiple time points is desired, initiate erasing of stored tension signals by the addition of an unlocking strand. To avoid excess rinsing, a higher final concentration of unlocking strand at 2 µM is used to initiate a toehold-mediated strand displacement reaction with the locking strand for 3 min, which erases the stored signals (Figure 3J). Gently rinse the excess oligonucleotides off with HBSS. The DNA hairpin tension probe surface and the cells are ready for another round of tension storing and mapping. Unlocking of tension signals is not necessary if only one time point is of interest in the study.
4. Data analysis
NOTE: Image analysis is performed using Fiji software, and the quantitative analysis is performed using analysis software.
- Correct any drift during image acquisition with Correct 3D drift command in Registration under the Plugins menu.
- Remove the camera background of the image with Subtract command under the Process menu.
- Determine the Pearson's correlation coefficient with the Colocalization function under Analyze menu.
- Average and subtract the fluorescence background produced by the unopened probes from three different local background regions. Draw ROIs of cells on either background-subtracted images or RICM (reflection interference contrast microscopy) images with Image J Freehand Selections tool. Measure any metric of interest of the ROIs, e.g., integrated fluorescence intensity and tension occupancy using Measure tool under Analyze menu (Figure 6).
- Export the measurements for quantitative analysis with analysis software.
- Plot the data with any analysis software.
Results
Here we show representative surface quality control images (Figure 4). A high-quality surface should have a clean background in RICM channel (Figure 4B), and uniform fluorescence intensity in Cy3B channel (Figure 4C). With the same imaging equipment and identical fluorescence imaging acquisition conditions, the background fluorescence intensity should be consistent and reproducible each time when conducting experiments with DNA prob...
Discussion
With the detailed procedures provided here, one can prepare DNA hairpin tension probe substrates to map and quantify the receptor tension produced by immune cells. When cells are plated onto the DNA hairpin tension probe substrate, they land, attach, and spread as the receptors sense the ligands both chemically and mechanically, the latter of which is detected by our probes. However, in some cases cells may fail to spread (Figure 7A) or fail to produce tension signal. This is often a consequ...
Disclosures
The authors declare no conflict of interest.
Acknowledgements
This work was supported by NIH Grants R01GM131099, NIH R01GM124472, and NSF CAREER 1350829. We thank the NIH Tetramer Facility for pMHC ligands. This study was supported, in part, by the Emory Comprehensive Glycomics Core.
Materials
| Name | Company | Catalog Number | Comments |
| 3-hydroxypicolinic acid (3-HPA) | Sigma | 56197 | maldi-TOF-MS matrix |
| mPEG-SC | Biochempeg | MF001023-2K | surface prep |
| (3-Aminopropyl)triethoxysilane | Acros | AC430941000 | surface prep |
| 10x Red blood cell lysis buffer | Biolegend | 00-4333-57 | buffer |
| 8.8 nm gold nanoparticles, tannic acid | Nanocomposix | customized order | surface prep |
| Atto647N NHS ester | Sigma | 18373-1MG-F | fluorophore, oligo prep |
| Attofluor Cell Chamber, for microscopy | Thermo Fisher Scientific | A7816 | imaging |
| BD Syringes only with Luer-Lok | BD bioscience | 309657 | cells |
| biotinylated anti-mouse CD3e | ebioscience | 13-0031-82 | antibody/ligand |
| Biotinylated pMHC ovalbumin (SIINFEKL) | NIH Tetramer Core Facility at Emory University | NA | antibody/ligand |
| bovine serum albumin | Sigma | 735078001 | block non-specific interactions |
| Cell strainers | Biologix | 15-1100 | cells |
| Coverslip Mini-Rack, teflon | Thermo Fisher Scientific | C14784 | surface prep |
| Cy3B NHS ester | GE Healthcare | PA63101 | fluorophore, oligo prep |
| Dulbecco's phosphate-buffered saline (DPBS) | Corning | 21-031-CM | buffer |
| ethanol | Sigma | 459836 | surface prep |
| Hank’s balanced salts (HBSS) | Sigma | H8264 | buffer |
| hydrogen peroxide | Sigma | H1009 | surface prep |
| LA-PEG-SC | Biochempeg | HE039023-3.4K | surface prep |
| Midi MACS (LS) startup kit | Miltenyi Biotec | 130-042-301 | cells |
| mouse CD8+ T cell isolation kit | Miltenyi Biotec | 130-104-075 | cells |
| Nanosep MF centrifugal devices | Pall laboratory | ODM02C35 | oligo prep |
| No. 2 round glass coverslips | VWR | 48382-085 | surface prep |
| NTA-SAM | Dojindo Molecular Technologies | N475-10 | surface prep |
| P2 gel | Bio-rad | 1504118 | oligo prep |
| sufuric acid | EMD Millipore Corporation | SX1244-6 | surface prep |
| Sulfo-NHS acetate | Thermo Fisher Scientific | 26777 | surface prep |
| Equipment | |||
| Agilent AdvanceBio Oligonucleotide C18 column, 4.6 x 150 mm, 2.7 μm | 653950-702 | oligonucleotide preparation | |
| Barnstead Nanopure water purifying system | Thermo Fisher | water | |
| CFI Apo 100× NA 1.49 objective | Nikon | Microscopy | |
| Cy5 cube | CHROMA | Microscopy | |
| evolve electron multiplying charge coupled device (EMCCD) | Photometrics | Microscopy | |
| High-performance liquid chromatography | Agilent 1100 | oligonucleotide preparation | |
| Intensilight epifluorescence source | Nikon | Microscopy | |
| Matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF-MS) | Voyager STR | oligonucleotide preparation | |
| Nanodrop 2000 UV-Vis Spectrophotometer | Thermo Fisher | oligonucleotide preparation | |
| Nikon Eclipse Ti inverted microscope | Nikon | Microscopy | |
| Nikon Perfect Focus System | Nikon | Microscopy | |
| NIS Elements software | Nikon | Microscopy | |
| quad band TIRF 405/488/561/647 cube | CHROMA | Microscopy | |
| RICM cube | CHROMA | Microscopy | |
| TIRF launcher with 488 nm (50 mW), 561 nm (50 mW), and 640 nm | Coherent | Microscopy | |
| TRITC cube | CHROMA | Microscopy | |
| oligo name | 5' modification / 3' modification | sequence (5' to 3') | Use |
| 15mer amine locking strand | 5' modification: no modification 3' modification: /3AmMO/ | AAA AAA CAT TTA TAC CCT ACC TA | locking real-time tension signal |
| 15mer Atto647N locking strand | 5' modification: Atto647N 3' modification: /3AmMO/ | AAA AAA CAT TTA TAC CCT ACC TA | locking real-time tension signal |
| 15mer non-fluoresccent locking strand | 5' modification: no modification 3' modification: no modification | A AAA AAC ATT TAT AC | locking real-time tension signal for quantitative analysis |
| 4.7 pN hairpin strand | 5' modification: no modification 3' modification: no modification | GTGAAATACCGCACAGATGCGT TTGTATAAATGTTTTTTTCATTTAT ACTTTAAGAGCGCCACGTAGCC CAGC | hairpin probe |
| amine ligand strand | 5' modification: /5AmMC6/ 3' modification: /3Bio/ | CGCATCTGTGCG GTA TTT CAC TTT | hairpin probe |
| BHQ2 anchor strand | 5' modification: /5ThiolMC6-D/ 3' modification: /3BHQ_2/ | TTTGCTGGGCTACGTGGCGCTCTT | hairpin probe |
| Cy3B ligand strand | 5' modification: Cy3B 3' modification: /3Bio/ | CGCATCTGTGCG GTA TTT CAC TTT | hairpin probe |
| unlocking strand | 5' modification: no modification 3' modification: no modification | TAG GTA GGG TAT AAA TGT TTT TTT C | unlocking accumulated tension signal |
References
- Dustin, M. L. T-cell activation through immunological synapses and kinapses. Immunological Reviews. 221 (1), 77-89 (2008).
- Spillane, K. M., Tolar, P. B cell antigen extraction is regulated by physical properties of antigen-presenting cells. Journal of Cell Biology. 216 (1), 217-230 (2017).
- Feng, Y., et al. Mechanosensing drives acuity of αβ T-cell recognition. Proceedings of the National Academy of Sciences. 114 (39), 8204-8213 (2017).
- Hong, J., et al. A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nature Immunology. 19 (12), 1379-1390 (2018).
- Basu, R., et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 165 (1), 100-110 (2016).
- Bashour, K. T., et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proceedings of the National Academy of Sciences. 111 (6), 2241-2246 (2014).
- Ma, V. P. Y., Salaita, K. DNA nanotechnology as an emerging tool to study mechanotransduction in living systems. Small. 15 (26), 1900961 (2019).
- Liu, Y., Yehl, K., Narui, Y., Salaita, K. Tension sensing nanoparticles for mechano-imaging at the living/nonliving interface. Journal of the American Chemical Society. 135 (14), 5320-5323 (2013).
- Glazier, R., et al. DNA mechanotechnology reveals that integrin receptors apply pN forces in podosomes on fluid substrates. Nature Communications. 10 (1), 1-13 (2019).
- Galior, K., Liu, Y., Yehl, K., Vivek, S., Salaita, K. Titin-based nanoparticle tension sensors map high-magnitude integrin forces within focal adhesions. Nano Letters. 16 (1), 341-348 (2016).
- Zhang, Y., Ge, C., Zhu, C., Salaita, K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nature Communications. 5, 5167 (2014).
- Liu, Y., et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proceedings of the National Academy of Sciences. 113 (20), 5610-5615 (2016).
- Zhang, Y., et al. Platelet integrins exhibit anisotropic mechanosensing and harness piconewton forces to mediate platelet aggregation. Proceedings of the National Academy of Sciences. 115 (2), 325-330 (2018).
- Huang, J., et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 464 (7290), 932-936 (2010).
- Ma, R., et al. DNA probes that store mechanical information reveal transient piconewton forces applied by T cells. Proceedings of the National Academy of Sciences. 116 (34), 16949-16954 (2019).
- Hui, E., et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 355 (6332), 1428-1433 (2017).
- Whitley, K. D., Comstock, M. J., Chemla, Y. R. Elasticity of the transition state for oligonucleotide hybridization. Nucleic Acids Research. 45 (2), 547-555 (2016).
- Brockman, J. M., et al. Live-cell super-resolved PAINT imaging of piconewton cellular traction forces. Nature Methods. 17 (10), 1018-1024 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved