A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Generating and Analyzing High-Parameter Histology Images with Histoflow Cytometry
In This Article
Summary
Described here is a method that can be used to image five or more fluorescent parameters by immunofluorescent microscopy. An analysis pipeline for extracting single cells from these images and conducting single-cell analysis through flow cytometry-like gating strategies is outlined, which can identify cell subsets in tissue sections.
Abstract
The usage of histology to investigate immune cell diversity in tissue sections such as those derived from the central nervous system (CNS) is critically limited by the number of fluorescent parameters that can be imaged at a single time. Most immune cell subsets have been defined using flow cytometry by using complex combinations of protein markers, often requiring four or more parameters to conclusively identify, which is beyond the capabilities of most conventional microscopes. As flow cytometry dissociates tissues and loses spatial information, there is a need for techniques that can retain spatial information while interrogating the roles of complex cell types. These issues are addressed here by creating a method for expanding the number of fluorescent parameters that can be imaged by collecting the signals of spectrally overlapping fluorophores and using spectral unmixing to separate the signals of each individual fluorophore. These images are then processed using an analysis pipeline to take high-parameter histology images and extract single cells from these images so that the unique fluorescent properties of each cell can be analyzed at a single-cell level. Using flow cytometry-like gating strategies, cells can then be profiled into subsets and mapped back onto the histology sections to not only quantify their abundance, but also establish how they interact with the tissue environment. Overall, the simplicity and potential of using histoflow cytometry to study complex immune populations in histology sections is demonstrated.
Introduction
Inflammation driven by cells of the immune system and glial cells can contribute to chronic disorders of the CNS where each population can promote the activity of the other1,2,3. Understanding how the immune system interacts with these elements of the CNS to promote CNS inflammation is currently a major topic of interest and has been greatly facilitated by high-parameter techniques such as single-cell RNA sequencing. Through single-cell RNA sequencing, we have discovered that there is extensive communication occurring between glial cells and the immune system in several CNS disorders4,5,6. Understanding how these interactions are affecting these disorders will be crucial to elucidating the biology of these diseases.
One issue with single-cell sequencing analyses is that these techniques require that the tissue be disrupted to obtain single cells or nuclei, resulting in a complete loss of spatial information. Knowing where a cell exists in a tissue is critical for understanding the cell's role in driving inflammation. For example, immune cells such as B cells can concentrate in the CNS during neuroinflammation; however, they rarely enter the CNS parenchyma and instead concentrate in CNS barriers7. Given their localization, it is unlikely that these cells contribute to CNS inflammation by physically interacting with glial cells in the CNS parenchyma, suggesting any interactions they may be having with glial cells would occur through secreted factors. Additionally, the pathology occurring in CNS disorders often has structure8,9 such that a cell's localization in the tissue could critically determine whether it is actively contributing to the disorder or is a bystander. Thus, the usage of spatial orientation to evaluate a cell's role in pathology is essential.
Studying cells in tissue has typically been accomplished by using immunohistochemistry or immune fluorescence microscopy. An issue with these techniques is that they typically can only image up to four parameters simultaneously. This is a major limitation to these techniques, as we know from flow cytometry and single-cell RNA sequencing analyses that many cell populations require two or more parameters for their identification; also, the number of parameters required typically increases when looking for specific subsets of a cell type10. Thus, it is impractical to use standard imaging techniques for studying how subsets of cells may be interacting within a tissue.
This issue has been partially overcome through newer high-parameter methods that can retain spatial information, such as spatial RNA sequencing11 and imaging mass cytometry12. While these techniques are valuable, they do have several issues, such as not being widely available, reducing three-dimensional data into two dimensions, and requiring considerable expertise to execute. Another technique known as sequential staining, wherein tissues are stained with one set of antibodies followed by inactivation of the previous set of antibodies before staining with another set of antibodies, can achieve high-parameter histology without the need for specialized equipment or expertise13,14,15. However, sequential staining can be extremely labor intensive and requires a large amount of microscopy time, which may be impractical for laboratories that do not own a personal microscope. Thus, there is a need for techniques that can expand the number of fluorescent parameters that can be imaged at one time on microscopes that are widely available and in a timely fashion.
Once the high-parameter data has been acquired, another issue arises: conventional image analysis methods are unlikely to successfully analyze the data. Techniques such as manual counting or thresholding are only viable if the analysis consists of a single parameter or if multiple markers have the same localization where only overlapping signals are counted. This limitation makes traditional analysis inadequate to work with high-parameter datasets. Successful analysis of these datasets has been achieved by segmenting single cells from histology images and then conducting flow cytometry-like gating strategies to identify cell types16,17. However, another issue that affects these analyses is that they only work for datasets wherein all the cells of interest are physically separated from one another, as these techniques do not employ methods that can accurately separate cells that are in physical contact. Thus, a newer method is required that can conduct single-cell analyses on histology sections even if the cells are in physical contact.
In this article, a simple protocol called histoflow cytometry is described that has previously been introduced18 that expands the number of fluorescent parameters that can be imaged simultaneously using widely available microscopes. This protocol works by staining tissues with spectrally overlapping dyes and then using spectral compensation to remove bleed-through from overlapping channels to obtain clear single stains. To facilitate the analysis of high-parameter histology images, a detailed analysis pipeline is described that extracts single cells from tissue sections for the purpose of sorting cells into distinct populations using flow cytometry-like gating strategies. This protocol works in tissues where cells are diffusely present and in tissues where cells are closely compacted together, making this technique versatile for the study of tissues like the CNS in both homeostasis and neuroinflammation. Histoflow cytometry is, therefore, a useful technique for studying interactions between complex cell types that require multiple cell markers to define cells while maintaining spatial information.
Protocol
This protocol does not cover sectioning tissues for histology; please see Jain et al.18 or19 for descriptions of how to section tissues for histology. This protocol can be used with any sectioned tissues on glass slides. This article uses inguinal lymph nodes isolated from an immunized animal as described previously18. The procedure and timeline for this protocol are summarized in Figure 1. The details of the reagents and the equipment used in this study are listed in the Table of Materials.
1. Staining tissue sections
NOTE: In addition to staining tissues with all of the antibody labels the researcher is interested in, the researcher should also prepare single color controls (one for each fluorescent parameter that is intended to be used) on adjacent or otherwise identical tissue slides wherein each single color control is stained with the reagents required to produce the signal of one fluorescent parameter.
- Prepare Perm and blocking buffer and Perm and staining buffer (see Table 1 for the composition).
- Thaw tissue sections on glass slides at room temperature and wipe off the condensation using a wipe. Use a hydrophobic marker to outline the border of the tissue(s) that need to be stained and let the marks dry.
- Rehydrate the tissues by soaking the tissue sections in phosphate-buffered saline (PBS) for 1 min. Shake off the PBS and use a wipe to remove excess PBS without directly touching the tissue.
- Fix the tissues by adding enough 4% paraformaldehyde (PFA) to cover the tissue completely. Incubate at room temperature for 20 min.
CAUTION: PFA can be toxic and should be handled within a fume hood.
NOTE: This step is only required for fresh frozen tissue. If the tissue is already fixed, then ignore this step. PFA fixation is described here, but the tissue can be fixed using other methods depending on the needs of the experiment. - Wash the tissues in a PBS bath for 1 min with gentle rocking, and then remove the slide from the bath. Replace the PBS and repeat the wash 3 more times.
- Shake off excess PBS and use a wipe to remove excess PBS without directly touching the tissue.
- Determine which secondary antibodies need to be used in step 1.12 of the protocol and the species of animals from which the secondary antibodies are derived. Fortify the Perm and blocking buffer by adding serum from these animals to the Perm and blocking buffer to a final concentration of 5% for each species.
NOTE: Usage of secondary antibodies helps amplify signal intensity. Thus, it is recommended that primary antibodies that produce weak signals should be paired with secondary antibodies to amplify their signals. If all of the antibody targets that are being stained are bright enough to not require secondary antibodies, steps 1.7-1.12 and step 1.15 can be skipped. - Block non-specific binding of antibodies by adding enough fortified Perm and blocking buffer to cover each tissue. Incubate the slides for 1-8 h at room temperature in a humidified chamber protected from light.
- Add unconjugated primary antibodies to the Perm and staining buffer at experimentally determined dilutions (the species or immunoglobulin isotypes of the primary antibodies should all be different and distinguishable by the secondary antibodies the researcher intends to use in step 1.12).
- Dump off the Perm and blocking buffer and use a wipe to remove excess buffer without directly touching the tissue. Add enough Perm and staining buffer with primary antibodies to the tissue to cover it and incubate the slide at 4 °C for 12-48 h.
NOTE: The researcher will need to determine the optimal time for antibody incubation. - Prepare blocking buffer and staining buffer (Table 1).
- Add secondary antibodies to the staining buffer prepared in step 1.11 at experimentally determined dilutions. The secondary antibodies used should target the same species or antibody isotypes that were used in step 1.9.
- Prepare the quenching buffer (as in Table 1).
NOTE: This step helps reduce autofluorescence from the tissue. If this is not needed, steps 1.13 and 1.14 can be skipped. - Wash the tissues as described in steps 1.5-1.6. Then, add enough quenching buffer to cover the tissues on the slides and incubate the slides at room temperature for 1-2 min.
- Wash the tissue as described in steps 1.5-1.6. Then, add the secondary antibodies diluted in the staining buffer prepared in step 1.12 to the tissues such that the tissues are completely covered in the staining buffer. Incubate slides at 4 °C for 2-8 h.
- Prepare the blocking solution by adding 50 µL of normal serum from whatever species of antibodies will be used in step 1.18 (5% of each species) to the blocking buffer prepared in step 1.11.
- Wash the tissues as described in steps 1.5-1.6. Then, add the blocking solution with normal serum prepared in step 1.16 to the tissues so that they are completely covered in staining buffer. Incubate slides at room temperature for 1-8 h.
- Prepare the staining solution by adding fluorophore-conjugated primary antibodies at experimentally determined dilutions to the staining buffer prepared in step 1.11. Also, add a nuclear stain, such as nuclear yellow, to the staining solution at an experimentally determined dilution.
NOTE: Steps 1.18-1.19 are only required if the researcher needs to use additional antibody labels that will not require secondary antibodies for signal amplification. If not using primary conjugated antibodies, nuclei stains can be added at step 1.12. - Shake off the blocking buffer and use a wipe to remove excess buffer without directly touching the tissue. Add enough staining solution prepared in step 1.18 to the tissue to cover it and incubate at 4 °C for 12-48 h.
- Wash the tissues as described in steps 1.5-1.6. Add 5 drops of mountant to each slide without introducing bubbles, and gently and slowly tip a glass coverslip over the tissue to cover the tissue completely in mountant and exclude all the air bubbles. Store the slides at 4 °C in the dark until they are ready to be imaged.
2. Imaging the tissue sections
NOTE: At this stage, the tissue sections are stained with all the antibodies of interest for all conditions that are intended to be imaged, and single-color controls are stained with one fluorescent parameter on each section. Imaging should be done on a microscope that has multiple detectors that can be tuned to detect specific ranges of light, and ideally, it should have access to as many laser lines for excitation as possible.
- Insert one of the fully stained slides into the microscope and set up the microscope to image the section using the objective that is intended to be used for collecting data.
- Based on the fluorophores the tissue is stained with, identify the excitation laser lines needed to stimulate those fluorophores and identify the emission ranges for each fluorophore that contain the greatest amount of signal.
NOTE: Online fluorescence spectra viewing tools (for example, see the Table of Materials) can be useful for identifying the optimal excitation wavelengths of fluorophores and identifying the optimal ranges for fluorophore detection. Fluorophores used for histoflow cytometry should be chosen such that each fluorophore differs in the optimal laser line that the fluorophore is stimulated by or differs by the optimal range of emission wavelengths. Fluorophores that have closer excitation/emission wavelengths will be more difficult to separate. The fluorophores used for this study are detailed in the results section. - Open Software 1 (see Table of Materials) in a confocal microscope and select the configuration tab at the top. Then select the hardware button on the left side and change the bit-depth to 16-bit.
NOTE: It is recommended to use higher bit depths, such as 16-bit data, for data collection, as this will improve the accuracy of the spectral compensation. Additionally, higher pixel densities, greater numbers for line averages, and more steps in the z-stack will improve the quality of the spectral separation. - Select the Acquire tab at the top of the screen and input the format setting to include the pixel density of the image (typically, higher resolutions produce better results).
- If available, select bidirectional X acquisition to speed up imaging time, provided this feature has been adequately calibrated.
- Pick a pinhole value for the confocal microscope (typically between 400-600 nm) where smaller values will typically produce higher quality histoflow cytometry results, although smaller pinhole values will make it more difficult to acquire images and may require additional imaging time or higher excitation intensities to produce images.
- Select the Show Sequential Scan Panel at the top of the left menu to open the sequential scan option. Use the '+' sign to set up additional sequences, where each sequence will contain one excitation wavelength that was decided upon in step 2.2.
- Turn on all lasers that are needed by clicking on the ON buttons associated with lasers of interest in the center of the screen to turn them into an ON state. Go into each specific sequence and increase the laser power in the center of the screen to whatever value is desired, where each sequence will have one excitation wavelength being used.
- Turn on as many detectors as needed at the bottom portion of the screen in each sequence to image all the fluorophores that will be imaged on that laser line. Adjust the ranges of the detectors by double-clicking on their ranges and inputting the upper and lower ranges of wavelengths that need to be collected.
NOTE: It is recommended that high-sensitivity detectors be used where possible, and ideally, all channels should be collected on the same types of detectors, although this is not required. - Press the live button, then focus on the sample. Use the Auto Scale button to the left of the image in the software to evaluate whether channels are adequately saturated with signal. The user should adjust the gain values and laser intensities in each sequence until the autoscaling does not reach the maximum value (65535 for 16-bit data) but is very close to saturation for all channels.
NOTE: As different fluorophores can differ in their brightness and different antibody targets can differ in their abundance, the sensitivity of the detectors will need to be adjusted to be of low sensitivity for bright signals and high sensitivity for dim signals if both fluorophores are stimulated by the same laser (the laser intensity cannot change in this scenario as both fluorophores would be affected). For best results, the fluorescence signal from each channel in the experiment should be balanced so that each channel has roughly equivalent amounts of signal, which will reduce spectral bleed-through into other channels and result in stronger signals. As different samples may differ in the abundance of the markers of interest being stained, it is recommended that the microscope settings be tested on multiple experimental conditions to confirm that bleaching is not occurring under any of the experimental conditions. - Once the microscope settings are finalized, load the microscope with the single color control slides and find representative staining for each fluorophore. Set up a Z-stack by focusing below the sample and hitting the begin button in the z stack menu, then focusing above the sample and hitting the end button.
- Choose the number of steps and the distance between steps (and use the same number of steps and parameters such as pixel densities and bit depth that will be used to image the fully stained slides), then press the start button.
NOTE: Ideally, these images should include the brightest signals from real staining that can be found on the slide. - After the single color controls have been imaged, proceed to use the same microscope settings to image all the fully stained samples.
3. Fluorescence compensation
- Open .lif files in Software 1 and click on the process tab.
- Choose the Dye Separation module and then choose the Automatic Dye Separation option. Select the Manual method for Dye Separation and choose no rescaling.
- Open one of the single-color control images. Manually inspect the single color controls for spectral bleed-through from one channel to the next by observing greyscaled windows for each channel for evidence of a signal in inappropriate channels. Use the Auto Scale button if needed to see weak bleed-through.
- Begin to remove bleed-through by manually inputting numbers (typically 0-1, where 0 represents no bleed-through and 1 represents 100% bleed-through) into the matrix on screen, then pressing apply to test whether the bleed-through is adequately removed. This should be repeated until the signals in all other channels except the channel of interest have been reduced to background levels.
NOTE: Overcompensation will lead to inaccurate results, so the fluorescence must be reduced to background levels but not below them. Overcompensated data will appear as one channel having a signal and other channels appearing to have black holes where the signal of interest is (Figure 2). - Once one of the single-color controls is completed, record the values in the matrix and then reset the matrix, and move to the next single-color control. Repeat this process for each single color control.
NOTE: The first dye will be in the Dye 1 row, and the amount of fluorescence that needs to be removed from Dye 1 will be in the adjacent columns. - Assemble all the values acquired from each control into one matrix and apply this matrix to a fully stained sample. If the separation looks accurate, apply the settings to all the images in the experiment, otherwise, re-evaluate the single color controls.
NOTE: Effective compensation should remove most of the bleed-through across channels, as summarized and shown in Figure 3. - Save all the compensated files into a separate .lif file.
4. Identifying nuclei and cells using ilastik
- Open the compensated .lif files in FIJI20. Enable viewing stacks in a hyperstack and select an image.
- (Optional) Use a selection tool such as the rectangle tool to include the portions of the image that will be analyzed and exclude areas that will not be analyzed. Crop the image using the image > crop function.
NOTE: Reducing the size of the image is useful in subsequent steps to reduce random-access memory (RAM) consumption and increase processing speed. - Save the image as a .tif file. Also, save the image as an hdf5 file by using the ilastik plugin for FIJI by clicking on Plugins > ilastik > Export hdf5. Save the file as "name of choice".h5 with 0 compression.
- Open Ilastik21 and make a pixel classification project. Use the Add New > Add Separate images function to add at least 3 representative images by going to add new input data.
- Right-click on one of the images, select Edit properties, and change the Display Mode to greyscale.
- On the left of the screen, select the feature selection tab. Click on Select features and select all features up to the highest sigma value that the program will allow you to use, then press OK.
- Now select the Training tab. Use the paintbrush tool (while label one is selected) to highlight individual cells (Figure 4) of interest such that the interior (cytoplasm and nucleus) and the cell membrane are included in the highlight.
- Repeat this across all the training samples where all cell types of interest are selected, and multiple cells of varying intensities, morphologies, and localizations in the tissue should be selected from each sample.
NOTE: Depending on the markers chosen for an individual set of stains, there may or may not be intracellular stains or cell membrane stains for the cells of interest. Whatever markers are used, ensure that the highlight includes the outmost stain on the cells of interest and includes the cell interior regardless of whether an intracellular stain is used. - Next, use label two to highlight everything that is not the cells of interest (Figure 4). This can include blank space, unlabeled nuclei, or tissue that does not have markers of interest. Ensure to include the space immediately adjacent to cells of interest to promote the learning of exact cell boundaries.
- Use the Live Update function to evaluate the quality of the training on images of interest. If training is inadequate, continue the training in steps 4.7 to 4.9. Otherwise, if satisfactory, click on the Prediction Export tab.
- Set the source to be probabilities. Click on the Choose Export Image Settings button, then set the c parameter to export 0-1 (assuming label 1 is highlighting parameters of interest).
- Convert the data type to a 16-bit Integer, and export as an hdf5 file to the directory of interest with the name of interest (we recommend using {nickname} mask.h5), then press OK.
- Click on the Batch Processing tab. Press the Select Raw Data files button, select all .h5 files of interest that require processing, and then press Open. Click on Process all files.
- Repeat steps 4.4 to 4.13, except instead of identifying cells, now identify the nuclei and exclude everything else (Figure 4). Export this as a separate prediction for identifying cell nuclei.
- Open Ilastik and create a new Data Conversion project. Add a representative hdf5 file into the Input data tab using the Add New > Add Separate Image function.
- Click on the Data Export tab and choose the Choose Export Image Settings button with the source set to input. Change the format to multipage tiff, choose a save location with the select function, and set the name of the file to {nickname}. Press OK.
- Click on the Batch processing tab and select all your hdf5 files using the Select Input Data Files button and press the Process all files button.
- Open Software 2 (see Table of Materials) and add all the tif/tiff files generated in steps 4.3 and 4.17 using the Add Files button. Choose a location for file outputs and press the Start All button to convert all of the files into .ims files. The files are now ready for analysis in Software 3 (see Table of Materials).
5. Analysis in Software 3
- Open one of the images containing all the fluorescent stains. Select Edit-> Add Channels and add the .ims files corresponding to the masks made over nuclei and cell bodies. Repeat this for all samples.
- Select the Add new Cells button. Select the detect nucleus and cell option and proceed forward.
- Select the mask over nuclei to be the source channel for detecting nuclei and select the advanced option for splitting nuclei by seed points. Choose a value for the nucleus diameter) and proceed forward.
NOTE: The optimal value for the nucleus diameter will need to be tested by running the simulation and manually evaluating whether the program is not splitting enough nuclei (multiple nuclei are grouped together) or whether there is over splitting (one nucleus is split into more than one). - Choose a value for the quality evaluation, and then proceed. Typically, it is best to include all nuclei, as the omission of nuclei can lead to multiple cells being fused into one.
- Choose a threshold value for creating nuclear surfaces and then proceed. Ensure that all nuclei, even ones that are dimmer, are still included in the threshold. Do not make the threshold too low, as this may overestimate the size of nuclei or potentially include debris as nuclear staining.
- Select a value for the number of minimum number Voxels per nucleus and proceed. Typically, it is best to keep everything unless debris or non-nuclei can be excluded using this parameter.
- Choose the Cell boundary and cytoplasm option, select the mask over cells to be the source channel for detecting cell bodies, and then proceed.
- Choose a Cell threshold that accurately captures all the cells of interest but is not so low that it includes background or any non-specific detection of cells. Select the option to split touching cells such that there is one nucleus per cell, and they are separated by the distance from nuclei. Also, choose to expand cells on nuclei and then proceed.
- Select a value for the minimum number of voxels per cell and proceed. Typically, it is best to keep everything unless debris or non-cells can be excluded using this parameter.
- Evaluate whether the created surfaces over the cells are accurate by looking at whether multiple cells have been fused together or if cells are missing. If the surfaces are inadequate, go back to step 5.2 and try new parameters until optimal settings are determined.
- Once the best settings are established, click on the created cells object in the left menu, click on the creation tab, and click the Store Parameters for Batch button to save the used settings.
- Open multiple windows of Software 3 and open separate images in each window. Press the new Cells button in each window, choose the saved settings in the Favorite Creation Parameters menu, and instruct each window to run till completion. Repeat this as many times as needed until all samples are analyzed.
NOTE: Running multiple instances of Software 3 will make this analysis proceed faster, but the number of windows that can be run in parallel will depend on the central processing unit (CPU) processing speed and the amount of RAM that the computer that the analysis is being run on. - Once all samples are completed, click on the created cells object in the left menu in each image and click on the Statistics tab. Click the Configure list of visible Statistics Values button, deselect all statistics except the 'Cell Intensity Mean' value, and then press OK.
- For every image that needs to be analyzed, click the Export all statistics to file button in the statistics tab and save the files to a location.
6. Histoflow cytometry analysis and mapping populations onto tissue sections
- Open Anaconda and JupyterLab within Anaconda. Next, open the script provided in Supplementary Coding File 1. Run the code using the Run menu and select Run All Cells, or run the cells individually in order by selecting Run Selected Cells.
- When prompted to input the source file, input the file directory for the exported fluorescent values generated in step 5.14. The file directory can be obtained by looking at the properties of one of the exported .csv files and copying the file path into the prompt.
- Next, the program will ask for an output location for the processed files. Input the file directory for any location that is suitable for saving the processed files. The program will automatically detect the number of fluorescent channels contained in the folder.
- Run the next section of the code to annotate the data with the names of each channel. This code can be modified as needed to change the "numberOfChannels" parameter to match the number of channels detected in step 6.3. Additionally, the names of the channels can be modified for as many channels as needed by repeating the 'ChX':'Marker' line as many times as needed and by modifying the names of the markers.
- Running the next line of code will create a .csv file at the specified location that is annotated and contains all the fluorescent parameters in the dataset. Repeat steps 6.1-6.5 for each set of Software 3 exports to create .csv files for all the data.
- Convert the .csv files to .fcs files by opening Software 4 (see Table of Materials) and dragging the .csv files into the program window. Software 4 will automatically start converting the files to .fcs files.
- Double-click on one of the samples in Software 4 to open a window on that sample. Use the gating tools in Software 4 to create a gating strategy. It is recommended to start by gating on a lineage-defining marker containing all the cells of interest, then proceeding to gates that remove rarer subsets first, and then finishing on the more abundant cell types.
- Once a gating strategy is established, test the accuracy of the gating by right-clicking on that population in the menu and choosing export. Export all parameters as a .csv file with headers included.
- Open JupyterLab in Anaconda. Next, open the script provided in Supplementary Coding File 2, and run the code. When prompted to input the location of the Software 4 file, input the file directory for the exported .csv file generated in the previous step. The file directory can be obtained by looking at the properties of the exported .csv file and copying the file path into the prompt.
- When prompted to add the output location, choose any file directory of choice, and ensure that the file directory includes the file name at the end ('file directory'\filename.txt). Run the rest of the code.
- Copy the text in the .txt file and open the corresponding .ims file in Software 3 for the same sample that the exported data is based upon. Click on the Cell object in the file, switch to the statistics tab, paste the text into the search bar, and start the search. All cells of interest will be highlighted.
- Manually inspect whether all the cells of interest were captured using the gating strategy. If the numbers of cells are underestimated or overestimated (examples shown in Figure 5), go back to step 6.7, adjust the gate, and re-evaluate whether the cells are accurately captured with the new gate. Repeat this with all the gates in the gating strategy and evaluate the gating on multiple samples.
- When all the gates are validated, export data from Software 4 using the table editor function and analyze the data using the preferred method of analysis.
NOTE: Cell populations can be mapped back onto the tissue sections in Software 3 by identifying the gated population as in steps 6.8-6.11 and then creating a surface based on the selected cells. This can be done for each gated population, as shown in Figure 6 and Figure 7.
Results
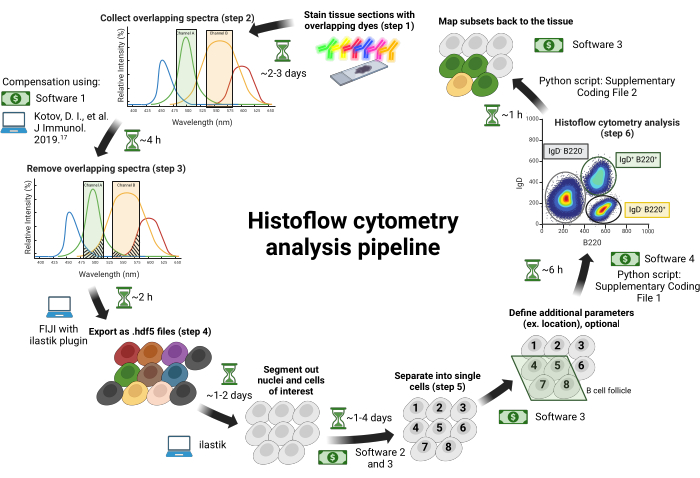
Figure 1: Histoflow cytometry workflow. Tissue sections are stained with spectrally overlapping dyes (step 1). Images are collected across individual excitation lasers paired with tunable bandpass filters to minimize spectral bleed-through between fluorophores (step 2). Spectral bleed-through between channels is corrected based on a compensatio...
Discussion
Here, the use of histoflow cytometry is described, a technique that has been validated previously18. It is demonstrated that when staining tissue sections with spectrally overlapping dyes, that bleed-through across channels can be removed using spectral compensation, resulting in a greater number of fluorescent parameters being clearly resolved than would normally be possible through conventional methods. As high-parameter histology images are difficult to analyze using conventional methods, an an...
Disclosures
The authors have no financial conflicts of interest.
Acknowledgements
We thank the Hotchkiss Brain Institute Advanced Microscopy Platform for imaging infrastructure and expertise. RWJ was supported by postdoctoral fellowship funding from the University of Calgary Eyes High program and by a Multiple Sclerosis Society of Canada and Roche Canada unrestricted educational fellowship. VWY received salary support from the Canada Research Chair Tier 1 program. This work was supported by operating funds from the Canadian Institutes of Health Research Grant 1049959, the Multiple Sclerosis Society of Canada Grant 3236, and the US Department of Defense of the Congressionally Directed Multiple Sclerosis Research Program. Figure 1 is created with BioRender.com. The figures adapted in this publication were originally published in The Journal of Immunology. Rajiv W. Jain, David A. Elliott, and V. Wee Yong. 2023. Single Cell Analysis of High-Parameter Histology Images Using Histoflow Cytometry. J. Immunol. 210: 2038-2049. Copyright © [2023]. The American Association of Immunologists, Inc.
Materials
| Name | Company | Catalog Number | Comments |
| 100% Ethanol | Sigma | 676829-1L | |
| 4% PFA | Electron Microscopy Sciences | 157-4 | |
| Anaconda | N/A | N/A | https://www.anaconda.com/download |
| Bovine Serum Albumin | Sigma | A4503-50G | |
| Cold fish stain gelatin | Sigma | G7765 | |
| Collating multichannel data from Imaris.ipynb script | N/A | N/A | https://github.com/elliottcalgary/Histoflow-Cytometry-Analysis- |
| Convert FlowJo output to txt file for Cell selection in Imaris.ipynb script | N/A | N/A | https://github.com/elliottcalgary/Histoflow-Cytometry-Analysis- |
| Donkey anti-rat Alexa Fluor 647 | JacksonImmunoResearch | 712-605-153 | 1:300 concentration |
| Donkey anti-rat DyLight 405 | Jackson ImmunoResearch | 712-475-153 | 1:200 concentration |
| Donkey Serum | JacksonImmunoResearch | 017-000-001 | |
| F(ab')2-Goat anti-Mouse IgG PerCP-eFluor 710 | Thermofisher | 46-4010-82 | 1:25 concentration |
| FIJI | N/A | N/A | https://imagej.net/software/fiji/ |
| FlowJo | FlowJo LLC | Software 4 | |
| Fluorescence spectraviewer | https://www.thermofisher.com/order/fluorescence-spectraviewer/#!/ | ||
| Fluoromount-G | Southern Biotech | 0100-01 | |
| Fresh frozen human tonsil sections | amsbio | HF-707 | |
| Glass coverslip | VWR | 48393 106 | |
| Goat anti-human IgA Alexa Fluor 488 | JacksonImmunoResearch | 109-546-011 | 1:400 concentration |
| Goat anti-human IgG Cy3 | JacksonImmunoResearch | 709-166-098 | 1:400 concentration |
| Goat anti-human IgM Dylight 405 | JacksonImmunoResearch | 109-476-129 | 1:300 concentration |
| Goat anti-rabbit A546 | Thermo Fisher Scientific | A-11035 | 1:250 concentration |
| Goat anti-rabbit IgG PE-Alexa Fluor 610 | Thermofisher | A-20981 | 1:250 concentration |
| Horse Serum | Sigma | H1138 | |
| Ilastik | N/A | N/A | https://www.ilastik.org/ |
| Ilastik FIJI plugin | N/A | N/A | https://www.ilastik.org/documentation/fiji_export/plugin |
| Imaris File Converter | Oxford Instruments | Software 2 | |
| Imaris with cell module | Oxford Instruments | Software 3 | |
| kimwipe | Kimtech | 34155 | |
| LasX Life Science software | Leica | Software 1 | |
| Mouse anti-human CD20 | VWR | CA95024-322 | 1:40 concentration |
| Mouse anti-human CD38 APC-R700 | BD Biosciences | 564980 | 1:20 concentration |
| Normal Goat Serum | JacksonImmunoResearch | 005-000-001 | |
| Normal Mouse Serum | JacksonImmunoResearch | 015-000-001 | |
| Normal Rabbit Serum | JacksonImmunoResearch | 011-000-001 | |
| Normal Rat Serum | JacksonImmunoResearch | 012-000-120 | |
| Nuclear Yellow | Abcam | ab138903 | Dissolve in DMSO at a concentration of 2 mg/ml and store at 4°C in the dark |
| PAP pen | Cedarlane | MU22 | |
| PBS | Gibco | 10010-023 | |
| Rabbit anti-human Ki67 | Abcam | ab15580 | 1:500 concentration |
| Rabbit anti-mouse Iba1 | Wako | 019-19741 | 1:500 concentration |
| Rat anti-human Blimp1 | Thermofisher | 14-5963-82 | 1:40 concentration |
| Rat anti-mouse B220 Alexa Fluor 647 | BioLegend | 103226 | 1:250 concentration |
| Rat anti-mouse CD138 | Biolegend | 142502 | 1:200 concentration |
| Rat anti-mouse CD3 PE-eFluor 610 | Thermo Fisher Scientific | 61-0032-82 | 1:40 concentration |
| Rat anti-mouse CD4 Alexa Fluor 488 | BioLegend | 100529 | 1:200 concentration |
| Rat anti-mouse CD45 allophycocyanin-R700 | BD Biosciences | 565478 | 1:50 concentration |
| Rat anti-mouse IgD PerCP-eFluor 710 | Thermo Fisher Scientific | 46-5993-82 | 1:50 concentration |
| SP8 Confocal microscope | Leica | ||
| Triton X-100 | Sigma | X100-500ml | |
| Trueblack | Biotium | 23007 | |
| Tween-20 | Sigma | P7949-500ml | |
| Ultracomp ebeads | Thermofisher | 01-2222-42 |
References
- Bar-Or, A., Li, R. Cellular immunology of relapsing multiple sclerosis: interactions, checks, and balances. Lancet Neurol. 20 (6), 470-483 (2021).
- Borst, K., Dumas, A. A., Prinz, M. Microglia: Immune and non-immune functions. Immunity. 54 (10), 2194-2208 (2021).
- Han, R. T., Kim, R. D., Molofsky, A. V., Liddelow, S. A. Astrocyte-immune cell interactions in physiology and pathology. Immunity. 54 (2), 211-224 (2021).
- Absinta, M., et al. A lymphocyte-microglia-astrocyte axis in chronic active multiple sclerosis. Nature. 597 (7878), 709-714 (2021).
- Piwecka, M., Rajewsky, N., Rybak-Wolf, A. Single-cell and spatial transcriptomics: Deciphering brain complexity in health and disease. Nat Rev Neurol. 19 (6), 346-362 (2023).
- Schirmer, L., et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 573 (7772), 75-82 (2019).
- Jain, R. W., Yong, V. W. B cells in central nervous system disease: Diversity, locations and pathophysiology. Nat Rev Immunol. 22 (8), 513-524 (2022).
- Lassmann, H. Multiple sclerosis pathology. Cold Spring Harb Perspect Med. 8 (3), (2018).
- Yong, V. W. Microglia in multiple sclerosis: Protectors turn destroyers. Neuron. 110 (21), 3534-3548 (2022).
- Sharma, S., Boyer, J., Teyton, L. A practitioner's view of spectral flow cytometry. Nat Methods. 21 (5), 740-743 (2024).
- Longo, S. K., Guo, M. G., Ji, A. L., Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. 22 (10), 627-644 (2021).
- Ramaglia, V., et al. Multiplexed imaging of immune cells in staged multiple sclerosis lesions by mass cytometry. Elife. 8, e48051 (2019).
- Bolognesi, M. M., et al. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J Histochem Cytochem. 65 (8), 431-444 (2017).
- Radtke, A. J., et al. IBEX: An iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues. Nat Protoc. 17 (2), 378-401 (2022).
- Radtke, A. J., et al. IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc Natl Acad Sci U S A. 117 (52), 33455-33465 (2020).
- Gerner, M. Y., Kastenmuller, W., Ifrim, I., Kabat, J., Germain, R. N. Histo-cytometry: A method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 37 (2), 364-376 (2012).
- Kotov, D. I., Pengo, T., Mitchell, J. S., Gastinger, M. J., Jenkins, M. K. Chrysalis: A new method for high-throughput histo-cytometry analysis of images and movies. J Immunol. 202 (1), 300-308 (2019).
- Jain, R. W., Elliott, D. A., Yong, V. W. Single-cell analysis of high-parameter histology images using histoflow cytometry. J Immunol. 210 (12), 2038-2049 (2023).
- JoVE Science Education Database. General Laboratory Techniques. Histological Sample Preparation for Light Microscopy. JoVE. , (2023).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 9 (7), 676-682 (2012).
- Berg, S., et al. ilastik: Interactive machine learning for (bio)image analysis. Nat Methods. 16 (12), 1226-1232 (2019).
- Reichard, A., Asosingh, K. Best practices for preparing a single cell suspension from solid tissues for flow cytometry. Cytometry A. 95 (2), 219-226 (2019).
- Ruhlandt, D., et al. Absolute quantum yield measurements of fluorescent proteins using a plasmonic nanocavity. Commun Biol. 3 (1), 627 (2020).
- Combs, C. A., Shroff, H. Fluorescence microscopy: A concise guide to current imaging methods. Curr Protoc Neurosci. 79, 1-25 (2017).
- Elliott, A. D. Confocal microscopy: Principles and modern practices. Curr Protoc Cytom. 92 (1), e68 (2020).
- Jonkman, J., Brown, C. M., Wright, G. D., Anderson, K. I., North, A. J. Tutorial: Guidance for quantitative confocal microscopy. Nat Protoc. 15 (5), 1585-1611 (2020).
- McNally, J. G., Karpova, T., Cooper, J., Conchello, J. A. Three-dimensional imaging by deconvolution microscopy. Methods. 19 (3), 373-385 (1999).
- Stringer, C., Wang, T., Michaelos, M., Pachitariu, M. Cellpose: A generalist algorithm for cellular segmentation. Nat Methods. 18 (1), 100-106 (2021).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved