Un abonnement à JoVE est nécessaire pour voir ce contenu. Connectez-vous ou commencez votre essai gratuit.
Method Article
Utilisation d’oligonucléotides synthétiques modifiés pour doser des enzymes métabolisant des acides nucléiques
Dans cet article
Résumé
Ici, un protocole pour doser les enzymes métabolisant les acides nucléiques est présenté, à l’aide d’exemples d’enzymes ligase, nucléase et polymérase. Le test utilise des oligonucléotides marqués par fluorescence et non marqués qui peuvent être combinés pour former des duplex imitant les dommages à l’ARN et/ou à l’ADN ou les intermédiaires de voie, permettant la caractérisation du comportement enzymatique.
Résumé
La disponibilité d’une gamme d’oligonucléotides synthétiques modifiés auprès de fournisseurs commerciaux a permis la mise au point de tests sophistiqués pour caractériser diverses propriétés des enzymes métabolisantes des acides nucléiques qui peuvent être exécutés dans n’importe quel laboratoire de biologie moléculaire standard. L’utilisation d’étiquettes fluorescentes a rendu ces méthodes accessibles aux chercheurs disposant d’un équipement d’électrophorèse PAGE standard et d’un imageur fluorescent, sans utiliser de matières radioactives ni nécessiter un laboratoire conçu pour le stockage et la préparation de matières radioactives, c’est-à-dire un laboratoire chaud. L’ajout facultatif de modifications standard telles que la phosphorylation peut simplifier la configuration du test, tandis que l’incorporation spécifique de nucléotides modifiés qui imitent les dommages à l’ADN ou les intermédiaires peut être utilisée pour sonder des aspects spécifiques du comportement enzymatique. Ici, la conception et l’exécution de tests pour interroger plusieurs aspects du traitement de l’ADN par des enzymes à l’aide d’oligonucléotides synthétiques disponibles dans le commerce sont démontrées. Il s’agit notamment de la capacité des ligases à se joindre ou des nucléases à dégrader différentes structures hybrides d’ADN et d’ARN, de l’utilisation différentielle de cofacteurs par l’ADN ligase et de l’évaluation de la capacité de liaison à l’ADN des enzymes. Les facteurs à prendre en compte lors de la conception de substrats nucléotidiques synthétiques sont discutés, et un ensemble de base d’oligonucléotides pouvant être utilisés pour une gamme de dosages de ligase d’acide nucléique, de polymérase et d’enzymes nucléases est fourni.
Introduction
Toutes les formes de vie ont besoin d’enzymes de traitement des acides nucléiques pour mener à bien les processus biologiques fondamentaux, notamment la réplication, la transcription et la réparation de l’ADN. Les principales fonctionnalités enzymatiques de ces voies sont les polymérases, qui génèrent des copies de molécules d’ARN/ADN, les ligases qui se joignent aux substrats polynucléotidiques, les nucléases qui les dégradent, et les hélicases et topoisomérases, qui font fondre les duplex d’acide nucléique ou modifient leur topologie 1,2,3,4,5,6,7,8,9,10 . De plus, bon nombre de ces enzymes fournissent des outils moléculaires essentiels pour des applications telles que le clonage, le diagnostic et le séquençage à haut débit 11,12,13,14,15.
Les caractéristiques fonctionnelles, la cinétique et les spécificités des substrats de ces enzymes peuvent être déterminées à l’aide de substrats d’ADN/ARN marqués produits par des oligonucléotides de recuit. Le suivi des substrats et des produits est traditionnellement réalisé par l’introduction d’un marqueur radioactif (32P) à l’extrémité du brin 5', qui peut ensuite être détecté par un film photographique ou à l’aide d’un système d’imagerie au phosphore16,17. Bien que les substrats radiomarqués offrent l’avantage d’une sensibilité expérimentale accrue et ne modifient pas les propriétés chimiques d’un nucléotide, les risques potentiels pour la santé liés au travail avec des radio-isotopes ont encouragé le développement d’un marquage des acides nucléiques non radioactifs afin de fournir une alternative plus sûre pour la détection de l’ADN et de l’ARN 18,19,20 . Parmi ceux-ci, la détection de fluorescence, y compris la détection de fluorescence directe, la fluorescence résolue dans le temps et les tests de transfert d’énergie/extinction de fluorescence se distinguent comme les plus polyvalents 21,22,23,24. La vaste gamme de fluorophores permet différentes conceptions de substrats d’ADN/ARN avec des rapporteurs uniques sur chaque oligonucléotide25. De plus, la stabilité des fluorophores, par rapport aux radio-isotopes, permet aux utilisateurs de produire et de préserver des quantités importantes de substrats d’ADN marqués par fluorescence19. Ces substrats marqués au fluorophore peuvent être incubés avec la protéine d’intérêt, ainsi que différentes combinaisons de cofacteurs métalliques et nucléotidiques, pour analyser la liaison et/ou l’activité enzymatique. La visualisation de la liaison ou de l’activité peut être observée à l’aide de divers canaux de colorant fluorophore avec un système d’imagerie sur gel. Comme seuls les oligonucléotides marqués par fluorescence seront visibles à l’aide de cette technique, toute augmentation ou diminution de la taille de l’oligonucléotide marqué sera facile à suivre. Les gels peuvent également être colorés par la suite, avec des colorants de coloration aux acides nucléiques pour visualiser toutes les bandes d’ADN présentes sur le gel.
Les ligases d’acides polynucléiques sont des enzymes qui joignent des fragments d’ADN/ARN, catalysant le scellement des cassures par la formation d’une liaison phosphodiester entre les terminaisons de l’ADN phosphorylé 5' et les 3' OH de l’ADN. Ils peuvent être divisés en deux groupes en fonction de leurs besoins en substrat nucléotidique. Les ligases dépendantes du NAD, hautement conservées, se trouvent dans toutes les bactéries26, tandis que les enzymes dépendantes de l’ATP structurellement diverses peuvent être identifiées dans tous les domaines de la vie 8,27. Les ligases d’ADN jouent un rôle important dans le traitement des fragments d’Okazaki pendant la réplication et sont impliquées dans diverses voies de réparation de l’ADN, telles que la réparation par excision de nucléotides et de bases, en scellant les entailles spontanées et les entailles laissées après la réparation 8,10. Différentes ligases d’ADN présentent des capacités variables pour joindre différentes conformations de cassures d’ADN, y compris des entailles dans un duplex, des cassures double brin, des discordances et des lacunes, ainsi que des hybrides d’ARN et d’ADN 28,29,30. Une gamme variée de substrats ligaturables peut être assemblée en recuit d’oligonucléotides avec un phosphate 5' pour générer des terminaisons 5' et 3' juxtaposées dans un duplex d’acide nucléique 31,32,33. La méthode d’analyse la plus courante est la résolution par PAGE d’urée dans un format de test final ; Cependant, les innovations récentes ont inclus l’utilisation de l’électrophorèse sur gel capillaire, qui permet un débit élevé34, le profilage par spectrométrie de masse35, ainsi qu’un test de balise moléculaire homogène, qui permet une surveillance résolue dans le temps36.
La première étape d’une réaction de ligature est l’adénylation de l’enzyme ligase par l’adénosine triphosphate (ATP) ou le nicotinamide adénine dinucléotide (NAD), ce qui donne un intermédiaire enzymatique covalent. La deuxième étape de la réaction est l’adénylation du substrat d’acide nucléique à l’extrémité 5' du site de l’entaille, qui est suivie de la ligature des brins d’entaille d’acide nucléique. De nombreuses enzymes ligases qui sont exprimées de manière recombinante dans E. coli sont purifiées sous la forme adénylée et, par conséquent, sont capables de ligaturer avec succès les acides nucléiques sans l’ajout d’un cofacteur nucléotidique. Il est donc difficile de déterminer le type particulier de cofacteur nucléotidique dont ils ont besoin pour la ligature des acides nucléiques. En plus de décrire des tests permettant d’évaluer l’activité de l’ADN ligase, une méthode permettant de déterminer de manière fiable l’utilisation des cofacteurs en désadénylant l’enzyme à l’aide de substrats non marqués est également présentée.
Les nucléases sont un groupe vaste et diversifié d’enzymes modifiant l’ADN/ARN et d’ARN catalytiques qui clivent les liaisons phosphodiester entre les acides nucléiques37. Les fonctionnalités de l’enzyme nucléase sont nécessaires à la réplication, à la réparation et au traitement de l’ARN et peuvent être classées en fonction de leur spécificité de sucre pour l’ADN, l’ARN ou les deux. Les endonucléases hydrolysent les liaisons phosphodiester à l’intérieur d’un brin d’ADN/ARN, tandis que les exonucléases hydrolysent les brins d’ADN/ARN un nucléotide à la fois à partir de l’extrémité 3' ou 5' et peuvent le faire à partir de l’extrémité 3' à 5' ou de l’extrémité 5' à 3' de l’ADN38.
Alors que de nombreuses protéines nucléases sont non spécifiques et peuvent être impliquées dans plusieurs processus, d’autres sont très spécifiques pour une séquence particulière ou des dommages à l’ADN 6,39,40. Les nucléases spécifiques à une séquence sont utilisées dans un large éventail d’applications biotechnologiques, telles que le clonage, la mutagénèse et l’édition du génome. Les nucléases populaires pour ces applications sont les nucléases de restriction41, les nucléases à doigts de zinc42, les nucléases effectrices de type activateur transcriptionnel et, plus récemment, les nucléases CRISPR43 guidées par l’ARN. Des nucléases spécifiques aux dommages ont récemment été identifiées, telles que la nucléase EndoMS, qui a une spécificité pour les mésappariements dans l’ADN grâce à son domaine nucléase de type RecB spécifique aux mésappariements 5,44. Historiquement, les essais d’activité des nucléases ont été effectués sous forme d’essais discontinus avec des substrats radiomarqués ; Cependant, en plus de leurs autres inconvénients, ceux-ci ne permettent pas d’identifier le site qui est coupé par une protéine nucléase, ce qui est possible lors de l’utilisation de substrats marqués par fluorescence45,46. Plus récemment, des tests de nucléases continues ont été mis au point et fonctionnent en utilisant différents colorants d’ADN qui interagissent avec l’ADN dans différents états ; par exemple, émettre un signal fluorescent plus élevé lors de l’interaction avec l’ADNdb qu’à l’état non lié, ou se lier spécifiquement à des ARN courts47. D’autres tests de nucléases continues utilisent des épingles à cheveux d’ADN avec un groupe fluorophore à l’extrémité 5' et un quencher à l’extrémité 3', de sorte que la fluorescence augmente à mesure que l’oligonucléotide est dégradé en raison d’une séparation du fluorophore et du quencher48. Bien que ces tests permettent de caractériser la cinétique des protéines dégradant l’ADN, ils nécessitent une connaissance préalable de la fonction et du substrat de l’enzyme et sont également limités aux enzymes qui modifient la conformation de l’ADN pour provoquer une différence dans la liaison du colorant. Pour cette raison, les tests de paramètres qui résolvent les produits nucléases individuels sont toujours souhaitables pour fournir un aperçu des modifications de l’ADN causées par l’activité des protéines.
Ici, une procédure détaillée est présentée pour la conception d’oligonucléotides d’ADN/ARN marqués par fluorescence qui peuvent être mélangés et appariés pour générer des substrats permettant de tester l’activité de nouvelles enzymes nucléases, polymérases et ligases. La validation de cet ensemble de base de séquences d’oligonucléotides simplifie la conception expérimentale et facilite le profilage économique d’un large éventail de fonctionnalités enzymatiques sans avoir besoin d’acheter un grand nombre de substrats sur mesure. Une procédure détaillée est fournie pour l’exécution d’un test enzymatique standard de traitement de l’ADN avec ces substrats, en utilisant l’exemple de l’activité de l’ADN ligase et des modifications pour le dosage et l’analyse des enzymes nucléases et polymérases sont décrites. De plus, un test modifié pour déterminer la spécificité du cofacteur de l’enzyme ADN ligase avec une grande précision est fourni, et des sondes à double marquage sont utilisées pour évaluer l’assemblage de ligatures à plusieurs composants. Enfin, des modifications au format de base de l’essai sont discutées pour permettre son utilisation pour déterminer les interactions protéine-ADN avec les mêmes substrats par l’essai de déplacement de mobilité électrophorétique (EMSA).
Protocole
1. Conception et achat d’oligonucléotides
REMARQUE : Concevez des oligonucléotides monobrin à assembler et à recuire dans les duplex souhaités. Un ou plusieurs des brins d’un duplex doivent porter une fraction fluorescente pour suivre le traitement des oligonucléotides par l’enzyme d’intérêt. Le tableau 1 présente un ensemble de base de séquences monocaténaires pouvant être assemblées pour une gamme d’activités.
- Incorporer les modifications spécifiques nécessaires pour l’enzyme d’intérêt, comme décrit ci-dessous.
- Pour les substrats d’ADN ligase (Figure 1) : Assemblez le substrat le plus simple à partir de trois oligonucléotides : un brin donneur phosphorylé 5' (NL2), un brin accepteur 5' marqué FAM (NL1) et un complément qui relie les deux (NL3).
- Assurez-vous que les brins fournissant l’extrémité 5' de l’entaille ligaturable sont phosphorylés avant l’assemblage du mélange maître de substrat à l’étape 2. Commandez ceci en tant que modification sur NL2 (comme indiqué dans le tableau 1) ou utilisez la phosphorylation enzymatique avec la polynucléotide kinase T4 après remise en suspension des oligonucléotides.
- Inclure la phosphorylation 5'-terminale de NL6 et NL8, qui comprennent le complément des cassures double brin illustrées à la figure 1A (NL6/NL7 et NL8/NL9) car elle ressemble le plus au substrat naturel produit à partir d’une endonucléase de restriction. Utilisez un substrat à double marquage pour déterminer l’étendue relative de la ligature pour les assemblages en plusieurs parties (voir étape 6).
- Modifier le brin du complément pour produire des décalages (NL10) et des lacunes (NL11).
REMARQUE : Des variations sur le substrat à entailles simples sont illustrées à la figure 1A. Il est possible d’utiliser d’autres séquences pour produire une gamme encore plus large de décalages ou d’écarts plus longs en variant la position soulignée. - Remplacez les oligonucléotides d’ARN par des oligonucléotides d’ADN.
REMARQUE : Des variations sur le substrat entaillé simple sont illustrées à la figure 1B. Une gamme plus large de duplex ADN/ARN peut être générée par des combinaisons supplémentaires de l’ensemble de base donné ici pour générer, par exemple, des cassures double brin contenant à la fois de l’ARN et de l’ADN. Un exemple de cette variation est donné à l’étape 6 ci-dessous, où une stratégie de double étiquette est utilisée.
- Pour les substrats d’ADN polymérase : Assemblez les oligonucléotides NL1 et NL3 énumérés dans le tableau 1 pour obtenir un test simple d’extension-amorce. Étudier d’autres aspects de l’activité de la polymérase en introduisant des modifications dans les brins NL1 (amorce) ou NL3 (matrice).
- Incorporer des analogues de base endommagés dans l’oligonucléotide NL3 avant la position 20 pour déterminer la capacité de contourner les lésions endommagées sur le brin matrice.
- Incorporer des analogues de base endommagés dans l’oligonucléotide NL1 à la position 20 pour déterminer la capacité d’extension d’une amorce endommagée.
- Utilisez RNL1 ou RNL3 dans le duplex pour étudier l’extension d’une amorce d’ARN ou l’utilisation d’une matrice d’ARN.
- Pour les substrats de nucléases (Figure 2) : Assembler les oligonucléotides pour obtenir une gamme non exhaustive de substrats double et simple brin (Figure 2Ai) ainsi qu’une gamme de jonctions à rabats et évasés (Figure 2Aii) et de substrats endommagés (Figure 2B).
- Pour sonder les activités des ribonucléases, remplacez de manière itérative NL1, NL2 et NL3 par RNL1, RNL2 et RNL3. Utilisez des versions d’ARN supplémentaires de HJ5 et HJ6 pour élargir davantage cet ensemble.
- Utilisez les oligonucléotides MD5, MD6 et MD9 qui ont une modification placée au centre qui imite les dommages oxydatifs, un intermédiaire de réparation abasique ou un produit de désamination (Figure 2B). Les substrats détecteront le clivage du brin à cette position. Étiquetez le brin NL3 du complément avec un fluorophore orthogonal tel que TAMRA pour détecter le clivage double brin (voir étape 6).
- Utilisez le marquage orthogonal du complément pour détecter la coupe double brin aux sites non appariés sur les brins de la sonde (NL5 et ND9) et du complément (MD10 et NL10).
- Pour les substrats d’ADN ligase (Figure 1) : Assemblez le substrat le plus simple à partir de trois oligonucléotides : un brin donneur phosphorylé 5' (NL2), un brin accepteur 5' marqué FAM (NL1) et un complément qui relie les deux (NL3).
- Commandez des oligonucléotides synthétiques incorporant des fluorophores pertinents et d’autres modifications auprès d’un fournisseur commercial.
REMARQUE : Une échelle de synthèse de 100 nM et une purification HPLC après la synthèse conviennent aux tests décrits.
2. Assemblage et recuit de duplex d’acide nucléique
- Mise en suspension et dilution des oligonucléotides
- Avant l’ouverture, centrifugez les oligonucléotides lyophilisés dans leurs tubes de 2 ml à pleine vitesse dans une centrifugeuse de paillasse pendant 2 à 5 minutes pour vous assurer que l’acide nucléique se trouve au fond du tube.
- Préparer une pâte mère de 100 μM en remettant en suspension les oligonucléotides dans un tampon TE (10 mM de tris(hydroxyméthyl)aminométhane (Tris), 1 mM d’acide éthylènediaminetétraacétique (EDTA)). Assurez-vous que les oligonucléotides sont complètement remis en suspension par un vortex doux répété et une brève centrifugation à pleine vitesse.
- Préparez une pâte de 10 μM en diluant une aliquote de matière mère avec un tampon TE. Diluer la pâte de 10 μM avec de l’eau ultrapure (eau MQ) pour préparer des matières de travail avec des concentrations de 0,5 μM, 0,7 μM ou 2,5 μM comme indiqué dans le tableau 2.
- Assemblage et recuit des mélanges maîtres de réaction
- Utilisez des matériaux de travail pour constituer les mélanges maîtres de réaction en utilisant les combinaisons fournies dans le tableau 2 et les volumes indiqués dans le tableau 3. Pour le test standard de ligase de l’ADN et la plupart des autres tests décrits ici, la composition tampon finale est de 50 mM de Tris pH 8,0, 50 mM de NaCl, 10 mM de dithiothréitol (DTT) avec 10 mM de Mg comme cation divalent.
- Recuit les oligonucléotides dans un tube de PCR ou de microcentrifugation en chauffant à 95 °C pendant 5 min à l’aide d’un bloc chauffant ou d’un thermocycleur. Laisser refroidir à température ambiante pendant 30 min (volumes <1 ml) à 1 h (volumes >1 ml). Pour les oligonucléotides plus longs (>40 nt), effectuez un refroidissement plus lent à l’aide d’un thermocycleur avec une rampe de descente de 95 °C à 25 °C pendant 45 min, ou faites flotter le tube contenant le mélange de recuit dans un bécher de 1 L d’eau bouillante et laissez-le refroidir à température ambiante pendant la nuit.
- Ajoutez des cofacteurs nucléotidiques et d’autres composants tampons sensibles à la chaleur au mélange maître après refroidissement à température ambiante. Utilisez le mélange réactionnel final directement pour le dosage par l’ajout d’enzyme (voir l’étape 3 ci-dessous) ou conservez-le à -20 °C pour une utilisation future.
3. Configuration standard du test
- Montage et lancement de la réaction de dosage
- Combinez 22,5 μL du mélange maître de substrat d’intérêt avec 2,5 μL d’ADN ligase ou d’une autre enzyme d’intérêt dans un tube PCR. Exécutez des réactions en double ou en triple, surtout si les résultats seront quantifiés.
- Inclure un témoin sans protéine (tampon seulement) dans les échantillons d’essai. N’incluez aucune commande de cofacteur à ce stade, si nécessaire.
REMARQUE : Les enzymes peuvent être stockées à -20 °C dans du glycérol à 50 % v/v, ce qui permet de les pipeter directement à partir de la solution. Assurez-vous que les solutions enzymatiques contenant du glycérol sont bien mélangées avant l’ajout, soit par pipetage pour mélanger, soit par vortex doux.
- Transférez immédiatement les réactions dans un appareil de PCR à 25 °C et incubez pendant 30 min. Variez la température et la durée en fonction des conditions optimales pour l’activité enzymatique.
- Tremper les réactions en ajoutant 5 μL de colorant de charge (formamide à 95 %, acide éthylènediaminetétraacétique (EDTA) 0,5 M, bleu de bromophénol) et incuber à 95 °C pendant 5 min.
4. Analyse des résultats d’analyse
- Préparez les gels Tris-Borate-EDTA (TBE)-Urea PAGE comme décrit ci-dessous.
- Préparez un bouillon de 20 % d’acrylamide, de 7 M d’urée et d’une solution de TBE x. Pour l’ensemble d’oligonucléotides décrit ici, utilisez la solution Acrylamide/Bis dans un rapport de 29:1 pour une résolution optimale.
- Pour un gel, combiner 10 ml d’acrylamide à 20 % et une solution d’urée à 7 M avec 100 μL d’APS (10 %) et 3 μL de tétraméthyléthylènediamine (TMED) et couler dans une roulette de gel.
- Une fois le gel solidifié, analysez les échantillons sur le gel d’urée TBE à 45 - 55 °C.
- Pré-exécutez le gel dans 1x tampon TBE pendant 30 min à 10 mA par gel avec chauffage externe.
- Éliminez l’excès d’urée dans les puits du gel en rinçant avec 1x TBE à l’aide d’une pipette de pâturage.
- Chargez 10 μL de chaque réaction et faites fonctionner à 10 mA pendant 1,0 à 1,5 h avec chauffage externe.
- Visualisez le gel sur l’imageur avec les bons réglages pour le fluorophore choisi. Pour la FAM, utilisez un ensemble de filtres qui donne une excitation/émission à 495/519 nm, qui est stocké en tant que préréglage dans la plupart des imageurs.
- Quantifiez l’intensité de la bande du produit et du substrat à l’aide d’un logiciel de traitement d’image avec l’imageur, ou d’un programme externe tel que ImageJ49,50 et calculez le pourcentage du produit à l’aide de la formule
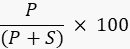
Où P est la valeur intégrée de la bande de produit et S est l’aire intégrée de la bande de substrat. Dans le cas de l’exemple de la réaction de ligase de l’ADN, la bande de produit est de 40 nucléotides (nt) et la bande de substrat de 20 nt.
5. Dé-adénylation de l’ADN ligase pour tester la spécificité du cofacteur
- Préparation de mélanges maîtres de réaction
- Préparez un ensemble du mélange maître contenant l’oligonucléotide NL1 marqué au FAM, comme décrit dans le tableau 4. Préparez une deuxième série contenant l’oligonucléotide NL1 sans étiquette FAM, comme décrit dans le tableau 4.
- Séparément, chauffer les deux duplex DNA à 95 °C pendant 5 min et refroidir pendant 30 min à 1 h à 25 °C. N’ajoutez pas de cofacteur nucléotidique à l’un ou l’autre des mélanges maîtres.
- Assemblage et initiation de la réaction de désadénylation
- Préparez une réaction de désadénylation unique pour chaque type/concentration de cofacteur à tester en combinant 10 μL du mélange maître non marqué avec 2,5 μL d’enzyme ligase.
- Préparez des tubes supplémentaires comme contrôle sans cofacteur et sans contrôle protéique (2,5 μL de tampon ajouté à la place de l’enzyme).
- Incuber les réactions à une température spécifique à l’activité optimale de l’enzyme, pendant 1 à 2 h. Le temps d’incubation peut être augmenté si l’enzyme est encore adénylée.
- Exécutez la réaction de ligature avec le cofacteur.
- Ajoutez 10 μL du mélange maître marqué et 2,5 μL des cofacteurs nucléotidiques souhaités (par exemple, ATP, NAD, ADP ou GTP) directement à la réaction désadénylée (concentration finale de 0,1 à 1 mM).
- Ajouter 2,5 μL de tampon de réaction au témoin sans cofacteur nucléotidique.
- Incuber les réactions pendant la même période et à la même température que celles utilisées précédemment. Trempez et visualisez comme décrit à l’étape 4.
6. Utilisation de substrats à double marquage pour la ligature par attelle ou l’assemblage en plusieurs parties
- Concevoir et acheter un oligonucléotide avec une fraction fluorescente qui a un spectre d’excitation/émission différent de celui du fluorophore déjà utilisé.
- Dans la configuration décrite ici, utilisez l’oligonucléotide NL2 (TAMRA) ayant de la 5-carboxytétraméthylrhodamine (TAMRA) à l’extrémité 3' (tableau 1).
- Assemblez le mixage principal comme décrit ci-dessous.
- Combinez les composants de la réaction décrits à l’étape 2, y compris les rapports équimolaires de tous les oligonucléotides utilisés dans l’assemblage, ainsi que les cations tampons et divalents.
- Recuit par chauffage à 95 °C pendant 5 min et refroidissement à 25 °C pendant 30 min - 1 h. Ajoutez le cofacteur et l’enzyme et incubez comme décrit à l’étape 3.
- Analysez et imagez les échantillons comme décrit à l’étape 4 en utilisant les canaux appropriés pour la paire de fluorophores dans le substrat. Dans le cas de FAM et TAMRA, il s’agit des canaux Fluorescéine (FITC) et Tétraméthylrhodamine (TRITC) présents sur la plupart des imageurs.
7. Évaluation de la liaison à l’ADN par Electrophoretic Mobility Shift Assay (EMSA) sur gel natif
- Préparez un gel TBE PAGE natif à 10 % comme décrit ci-dessous.
- Mélanger 2,5 ml d’acrylamide à 40 %, 1 mL de 10x TBE, 100 μL d’APS à 10 %, 3 μL de TMED et 6,5 mL d’eau MQ et couler dans un fondeur de gel.
- Assemblez la réaction de liaison comme décrit ci-dessous.
- Assemblez le substrat de l’EMSA conformément au tableau 5 de manière à ce que l’EDTA (10 mM) soit inclus et que les ions métalliques soient omis.
- Combinez 20 μL du mélange maître de substrat EMSA avec 5 μL de protéine dans un tube PCR. Inclure un échantillon de contrôle sans protéines. Incuber pendant 30 min à 25 °C.
- Analyser par électrophorèse native comme décrit ci-dessous.
- Ajouter 5 μL de colorant de charge natif 5x (100 mM d’EDTA, 0,25 % de bleu de bromophénol, 25 % de glycérol v/v et de l’eau MQ jusqu’à 1 mL) aux échantillons.
- Chargez sur le gel préparé et faites-le fonctionner à 60 V pendant 2-3 h avec refroidissement par circulation d’eau jusqu’à ce que le front du colorant soit à quelques cm au-dessus de l’extrémité du gel.
- Visualisez et analysez les gels comme décrit à l’étape 4.
Résultats
Ligature par l’ADN ligase
L’activité enzymatique de l’ADN ligase entraînera une augmentation de la taille de l’oligonucléotide marqué par fluorescence lorsqu’elle est visualisée sur un gel d’urée PAGE. Dans le cas des substrats pour la ligature de l’ADN et de l’ARN énumérés dans le tableau 2, cela correspond à un doublement de la taille de 20 nt à 40 nt (figure 3A). L’activité enzymatique optimale peut être déterminée par ...
Discussion
Étapes critiques du protocole
Conception et achat d’oligonucléotides : Lors de l’achat d’oligonucléotides pour la formation duplex, il est essentiel de prendre en compte la conception de séquences. Il est recommandé d’utiliser un outil d’analyse d’oligonucléotides pour prédire les propriétés de la séquence nucléotidique, telles que la teneur en GC, la température de fusion, la structure secondaire et le potentiel de dimérisation, avant d’en commander57
Déclarations de divulgation
SEG et UR sont des employés d’ArcticZymes Technologies AS qui distribue la R2D ligase. AW, ER-S et RS n’ont pas d’intérêts concurrents.
Remerciements
AW est soutenu par une bourse de découverte Rutherford (20-UOW-004). RS est le récipiendaire d’une bourse d’études de la Nouvelle-Zélande après l’Antarctique. SG et UR remercient l’Institut de chimie de l’Université de Tromsø - l’Université norvégienne de l’Arctique pour son soutien technique.
matériels
| Name | Company | Catalog Number | Comments |
| 30% Acrylamide/Bis Solution (29:1) | BioRad | 1610156 | |
| Adenosine triphosphate (ATP) | Many suppliers | ||
| Ammonium persulfate (APS) | Many suppliers | ||
| Benchtop centrifuge | Many suppliers | ||
| Borate | Many suppliers | ||
| Bromophenol blue | Many suppliers | ||
| Dithiothreitol (DTT) | Many suppliers | ||
| Electrophoresis system with circulating water bath | Many suppliers | ||
| Ethylenediaminetetraacetic acid (EDTA) | Many suppliers | ||
| Fluoresnence imager, e.g. iBright FL1000 | Thermo Fisher Scientific | A32752 | |
| Formamide | Many suppliers | ||
| Gel casting system | Many suppliers | ||
| Heating block | Many suppliers | ||
| Magnesium Chloride | Many suppliers | Other metal ions may be preferred depending on the protein studied | |
| Microcentrifuge tubes (1.5 mL) | Many suppliers | ||
| Micropipettes and tips | Many suppliers | 1 mL, 0.2 mL, 0.02 mL, 0.002 mL | |
| Nicotinamide adenine dinucleotide (NAD+) | Many suppliers | ||
| Oligonucleotides | Integrated DNA Technologies | NA | Thermo Fisher, Sigma-Aldrich, Genscript and others also supply these |
| pasture pipette | Many suppliers | ||
| PCR thermocycler | Many suppliers | ||
| PCR tubes | Many suppliers | ||
| RNAse away | ThermoFisher | 7002PK | Only needed when working with RNA oligos |
| RNase AWAY | Merck | 83931-250ML | Surfactant for removal of RNAse contamination on surfaces |
| RNAse-free water | New England Biolabs | B1500L | Only needed when working with RNA oligos |
| Sodium Chloride | Many suppliers | ||
| SUPERase IN RNase inhibitor | Thermo Fisher Scientific | AM2694 | Broad spectrum RNAse inhibitir (protein-based) |
| SYBR Gold | Thermo Fisher Scientific | S11494 | This may be used to post-stain gels and visualise unlabelled oligonucleotides |
| Tetramethylethylenediamine (TMED) | Many suppliers | ||
| Tris, or tris(hydroxymethyl)aminomethane | Many suppliers | ||
| Ultrapure water (Milli-Q) | Merck | ||
| urea | Many suppliers | ||
| Vortex | Many suppliers |
Références
- Gao, Y., et al. Structures and operating principles of the replisome. Science. 363 (6429), 7003 (2019).
- Yang, W., Gao, Y. Translesion and repair DNA polymerases: Diverse structure and mechanism. Annu Rev Biochem. 87, 239-261 (2018).
- Lohman, T. M., Fazio, N. T. How does a helicase unwind DNA? Insights from RecBCD Helicase. Bioessays. 40 (6), e1800009 (2018).
- Ahdash, Z., et al. Mechanistic insight into the assembly of the HerA-NurA helicase-nuclease DNA end resection complex. Nucleic Acids Res. 45 (20), 12025-12038 (2017).
- Wozniak, K. J., Simmons, L. A. Bacterial DNA excision repair pathways. Nat Rev Microbiol. 20 (8), 465-477 (2022).
- Zhang, L., Jiang, D., Wu, M., Yang, Z., Oger, P. M. New insights into DNA repair revealed by NucS endonucleases from hyperthermophilic Archaea. Front Microbiol. 11, 1263 (2020).
- Saathoff, J. H., Kashammer, L., Lammens, K., Byrne, R. T., Hopfner, K. P. The bacterial Mre11-Rad50 homolog SbcCD cleaves opposing strands of DNA by two chemically distinct nuclease reactions. Nucleic Acids Res. 46 (21), 11303-11314 (2018).
- Williamson, A., Leiros, H. S. Structural insight into DNA joining: from conserved mechanisms to diverse scaffolds. Nucleic Acids Res. 48 (15), 8225-8242 (2020).
- Caglayan, M. Interplay between DNA polymerases and DNA ligases: Influence on substrate channeling and the fidelity of DNA ligation. J Mol Biol. 431 (11), 2068-2081 (2019).
- Shuman, S. DNA ligases: Progress and prospects. J Biol Chem. 284 (26), 17365-17369 (2009).
- Lohman, G. J., Tabor, S., Nichols, N. M. DNA ligases. Curr Prot Mol Biol. , (2011).
- Rittié, L., Perbal, B. Enzymes used in molecular biology: a useful guide. J Cell Commun Signal. 2 (1-2), 25-45 (2008).
- Chandrasegaran, S., Carroll, D. Origins of programmable nucleases for genome engineering. J Mol Biol. 428 (5), 963-989 (2016).
- Aschenbrenner, J., Marx, A. DNA polymerases and biotechnological applications. Curr Opin Biotechnol. 48, 187-195 (2017).
- Loenen, W. A. M., Dryden, D. T. F., Raleigh, E. A., Wilson, G. G., Murray, N. E. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 42 (1), 3-19 (2013).
- Voytas, D., Ke, N. Detection and quantitation of radiolabeled proteins and DNA in gels and blots. Curr Protoc Immunol. , (2002).
- Phillips, D. H. Detection of DNA modifications by the 32P-postlabelling assay. Mutat Res. 378 (1-2), 1-12 (1997).
- Huang, C., Yu, Y. T. Synthesis and labeling of RNA in vitro. Curr Prot Mol Biol. , (2013).
- Ballal, R., Cheema, A., Ahmad, W., Rosen, E. M., Saha, T. Fluorescent oligonucleotides can serve as suitable alternatives to radiolabeled oligonucleotides. J Biomol Tech. 20 (4), 190-194 (2009).
- Anderson, B. J., Larkin, C., Guja, K., Schildbach, J. F. Using fluorophore-labeled oligonucleotides to measure affinities of protein-DNA interactions. Meth Enzymol. 450, 253-272 (2008).
- Liu, W., et al. Establishment of an accurate and fast detection method using molecular beacons in loop-mediated isothermal amplification assay. Sci Rep. 7 (1), 40125 (2017).
- Ma, C., et al. Simultaneous detection of kinase and phosphatase activities of polynucleotide kinase using molecular beacon probes. Anal Biochem. 443 (2), 166-168 (2013).
- Li, J., Cao, Z. C., Tang, Z., Wang, K., Tan, W. Molecular beacons for protein-DNA interaction studies. Meth Mol Biol. 429, 209-224 (2008).
- Yang, C. J., Li, J. J., Tan, W. Using molecular beacons for sensitive fluorescence assays of the enzymatic cleavage of nucleic acids. Meth Mol Biol. 335, 71-81 (2006).
- Nikiforov, T. T., Roman, S. Fluorogenic DNA ligase and base excision repair enzyme assays using substrates labeled with single fluorophores. Anal Biochem. 477, 69-77 (2015).
- Pergolizzi, G., Wagner, G. K., Bowater, R. P. Biochemical and structural characterisation of DNA ligases from bacteria and Archaea. Biosci Rep. 36 (5), 00391 (2016).
- Martin, I. V., MacNeill, S. A. ATP-dependent DNA ligases. Genome Biol. 3 (4), (2002).
- Bilotti, K., et al. Mismatch discrimination and sequence bias during end-joining by DNA ligases. Nucleic Acids Res. 50 (8), 4647-4658 (2022).
- Bauer, R. J., et al. Comparative analysis of the end-joining activity of several DNA ligases. PLoS One. 12 (12), e0190062 (2017).
- Lohman, G. J. S., Zhang, Y., Zhelkovsky, A. M., Cantor, E. J., Evans, T. C. Efficient DNA ligation in DNA-RNA hybrid helices by Chlorella virus DNA ligase. Nucleic Acids Res. 42 (3), 1831-1844 (2014).
- Bullard, D. R., Bowater, R. P. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem J. 398 (1), 135-144 (2006).
- Magnet, S., Blanchard, J. S. Mechanistic and kinetic study of the ATP-dependent DNA ligase of Neisseria meningitidis. Biochemistry. 43 (3), 710-717 (2004).
- Williamson, A., Grgic, M., Leiros, H. S. DNA binding with a minimal scaffold: structure-function analysis of Lig E DNA ligases. Nucleic Acids Res. 46 (16), 8616-8629 (2018).
- Lohman, G. J., et al. A high-throughput assay for the comprehensive profiling of DNA ligase fidelity. Nucleic Acids Res. 44 (2), e14 (2016).
- Kim, J., Mrksich, M. Profiling the selectivity of DNA ligases in an array format with mass spectrometry. Nucleic Acids Res. 38 (1), e2 (2010).
- Tang, Z. W., et al. Real-time monitoring of nucleic acid ligation in homogenous solutions using molecular beacons. Nucleic Acids Res. 31 (23), e148 (2003).
- Yang, W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 44 (1), 1-93 (2011).
- Marti, T. M., Fleck, O. DNA repair nucleases. Cell Mol Life Sci. 61 (3), 336-354 (2004).
- Wang, B. B., et al. Review of DNA repair enzymes in bacteria: With a major focus on AddAB and RecBCD. DNA Repair. 118, 103389 (2022).
- Pidugu, L. S., et al. Structural insights into the mechanism of base excision by MBD4. J Mol Biol. 433 (15), 167097 (2021).
- Roberts, R. J. How restriction enzymes became the workhorses of molecular biology. Proc Natl Acad Sci U S A. 102 (17), 5905-5908 (2005).
- Miller, J. C., et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 25 (7), 778-785 (2007).
- Kim, H., Kim, J. S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 15 (5), 321-334 (2014).
- Takemoto, N., Numata, I., Su'etsugu, M., Miyoshi-Akiyama, T. Bacterial EndoMS/NucS acts as a clamp-mediated mismatch endonuclease to prevent asymmetric accumulation of replication errors. Nucleic Acids Res. 46 (12), 6152-6165 (2018).
- Reardon, J. T., Sancar, A. Molecular anatomy of the human excision nuclease assembled at sites of DNA damage. Mol Cell Biol. 22 (16), 5938-5945 (2002).
- Kunkel, T. A., Soni, A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-gamma. J Biol Chem. 263 (9), 4450-4459 (1988).
- Sheppard, E. C., Rogers, S., Harmer, N. J., Chahwan, R. A universal fluorescence-based toolkit for real-time quantification of DNA and RNA nuclease activity. Sci Rep. 9 (1), 8853 (2019).
- Li, J. J., Geyer, R., Tan, W. Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res. 28 (11), e52 (2000).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 9 (7), 671-675 (2012).
- Sharma, J. K., et al. Methods for competitive enrichment and evaluation of superior DNA ligases. Meth Enzymol. 644, 209-225 (2020).
- Rzoska-Smith, E., Stelzer, R., Monterio, M., Cary, S. C., Williamson, A. DNA repair enzymes of the Antarctic Dry Valley metagenome. Front Microbiol. 14, 1156817 (2023).
- Williamson, A., Pedersen, H. Recombinant expression and purification of an ATP-dependent DNA ligase from Aliivibrio salmonicida. Protein Expres Purif. 97, 29-36 (2014).
- Akey, D., et al. Crystal structure and nonhomologous end-joining function of the ligase component of Mycobacterium DNA ligase D. J Biol Chem. 281 (19), 13412-13423 (2006).
- Kim, D. J., et al. ATP-dependent DNA ligase from Archaeoglobus fulgidus displays a tightly closed conformation. Acta Crystallogr Sect F Struct Biol Cryst Commun. 65, 544-550 (2009).
- Nishida, H., Kiyonari, S., Ishino, Y., Morikawa, K. The closed structure of an archaeal DNA ligase from Pyrococcus furiosus. J Mol Biol. 360 (5), 956-967 (2006).
- Gundesø, S., et al. R2D ligase: Unveiling a novel DNA ligase with surprising DNA-to-RNA ligation activity. Biotechnol J. 19 (3), e2300711 (2024).
- Hendling, M., Barišić, I. In silico design of DNA oligonucleotides: Challenges and approaches. Comput Struct Biotechnol J. 17, 1056-1065 (2019).
- Green, M. R., Sambrook, J. How to win the battle with RNase. Cold Spring Harb Prot. 2019 (2), (2019).
- Summer, H., Grämer, R., Dröge, P. Denaturing urea polyacrylamide gel electrophoresis (Urea PAGE). J Vis Exp. (32), e1485 (2009).
- Smith, D. R. Gel Electrophoresis of DNA. Mol Biometh Handbook. , 17-33 (1998).
- Lorenz, T. C. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. J Vis Exp. (63), e3998 (2012).
- Rousseau, M., et al. Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus. Nucleic Acids Res. 52 (7), 3924-3937 (2024).
- Kestemont, D., Herdewijn, P., Renders, M. Enzymatic synthesis of backbone-modified oligonucleotides using T4 DNA ligase. Curr Prot Chem Biol. 11 (2), e62 (2019).
- Farell, E. M., Alexandre, G. Bovine serum albumin further enhances the effects of organic solvents on increased yield of polymerase chain reaction of GC-rich templates. BMC Res Notes. 5, 257 (2012).
- Nazarenko, I., Pires, R., Lowe, B., Obaidy, M., Rashtchian, A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 30 (9), 2089-2195 (2002).
Réimpressions et Autorisations
Demande d’autorisation pour utiliser le texte ou les figures de cet article JoVE
Demande d’autorisationThis article has been published
Video Coming Soon