A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Measurement of Microtubule Dynamics by Spinning Disk Microscopy in Monopolar Mitotic Spindles
In This Article
Summary
Here we present a robust and detailed method of microtubule dynamics analysis in cells synchronized in prometaphase using live-cell spinning disk confocal microscopy and MATLAB-based image processing.
Abstract
We describe a modification of an established method to determine microtubule dynamics in living cells. The protocol is based on the expression of a genetically encoded marker for the positive ends of microtubules (EB3 labelled with tdTomato fluorescent protein) and high-speed, high-resolution, live-cell imaging using spinning disk confocal microscopy. Cell cycle synchronization and increased density of microtubules are achieved by inhibiting centrosomal separation in mitotic cells, and analysis of growth is performed using open-source U-Track software. The use of a bright and red-shifted fluorescent protein, in combination with the lower laser power and reduced exposure time required for spinning disk microscopy reduce phototoxicity and the probability of light-induced artifacts. This allows for imaging a larger number of cells in the same preparation while maintaining the cells in a growth medium under standard culture conditions. Because the analysis is performed in a supervised automatic fashion, the results are statistically robust and reproducible.
Introduction
Microtubules (MTs) are highly dynamic structures found in virtually all eukaryotic cells and in some bacteria1. Together with actin and intermediate filaments, they sculpt the cytoskeleton2,3. Cell division4, molecule transport5, flagellar beating6, the sensation of the surrounding environment through primary cilium7, hearing (kinocilium)8,9, embryogenesis10,11,12, invasion and metastasis13,14, and even memory formation15,16,17,18, and many other processes primarily rely on MTs. Participation of MTs in all these events would be impossible without their remarkable ability to rapidly switch between growth (polymerization) and shrinkage (depolymerization). This property is described as dynamic instability19. MT dynamicity is altered in many pathological conditions20,21,22. Hence, determining the nature of this property can help to understand disease mechanisms and subsequently their treatment.
A long list of methods has been developed for MT dynamics analysis, most of which are based on imaging techniques23. Initially, wide field light microscopes were used for observing the formation of tubulin polymers in vitro24. The discovery of end-binding (EB)-proteins that collect at MT plus-ends and the development of methods to fluorescently label proteins made it possible to observe the behavior of MTs directly in living cells with wide field and confocal fluorescence microscopes25,26,27. One EB-protein is end-binding protein 3 (EB3)28; by overexpressing and tracking EB3 fused to a fluorescent protein, MT plus-end assembly rates can be determined29,30.
Confocal laser scanning fluorescence microscopy (CLSM) is frequently used to follow MT dynamics. However, this imaging technique poses a high risk of phototoxicity and photobleaching, two undesirable processes for live cell and dim sample imaging31. In order to obtain a better signal-to-noise ratio, the laser power and the exposure duration should be high enough while not damaging the samples, and this requires sacrificing resolution in exchange for speed. A suitable alternative to CLSM is spinning disk microscopy32. This imaging modality is based on the use of a Nipkow disk33, which consists of a moving disk bearing an array of pinholes, and works equivalently to many CLS microscopes imaging the same sample simultaneously34. Therefore, the light from the laser will illuminate several regions in the sample simultaneously but retain the confocal nature. The Nipkow disk, therefore, allows obtaining images similar to CLSM but faster and using less laser power. The Nipkow disk was further improved by Yokogawa Electric, which introduced a second disk with an array of microlenses on it that individually direct light into a respective pinhole, further reducing phototoxicity and photobleaching35. Thus, spinning disk laser scanning microscopy became a method of choice for live cell imaging, and it makes it possible to obtain images with high signal-to-noise ratio at a high speed31,36, which is crucial for resolving signals such as those from the fast-growing MT ends.
MT dynamics differ temporarily. For example, the mitotic MTs are more dynamic than the interphase ones37,38. Similarly, differences in the growth rate and shrinkage have been observed even within the same cell cycle phase, such as mitosis39,40. Therefore, to avoid false data collection, the measurement of MT dynamics should be limited to a narrow time-window during the cell cycle. For example, measurement of MT dynamics in prometaphase can be achieved by treating the cells with dimethylenastron (DME), a monastrol analogue that inhibits the motor kinesin Eg541 and prevents the formation of the bipolar mitotic spindle42. Inhibition of cells at prometaphase with Eg5 inhibitor DME and other monastrol derivatives does not affect the MT dynamics43,44,45, which makes DME a useful tool for studying MT dynamics both in fixed and live cells44.
Here we combine the method of MT dynamics analysis in prometaphase cells described by Ertych et al.44 with dual spinning disk imaging. This method allows measurement of the MT dynamics in prometaphase cells collected from a single focal plane with a higher imaging rate, yet without photobleaching and minimal phototoxicity. Furthermore, as a fluorescent reporter, we use tandem dimer Tomato fluorescent protein (tdTomato) which has improved brightness and photostability in comparison to the green fluorescent protein (EGFP) and is excited with lower energy light46. Therefore, tdTomato requires less laser power for excitation and is less phototoxic. Altogether, we further improve the method by reducing the phototoxicity and improving the resolution and postprocessing required for the MT dynamics analysis. Additionally, we create a basis for future modifications of the method by combining it with other synchronization techniques.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Seeding of HeLa Cells
- Prepare 2 mL of 5 μg/mL fibronectin solution in phosphate buffered saline (PBS) and add 450 μL of it into each well of a 4 well chambered coverslip (#1.5). Incubate the slide for 15 min at 37 °C and 5% CO2.
- Rinse asynchronously growing HeLa cells with Dulbecco’s Phosphate Buffered Saline (DPBS) and incubate with trypsin-EDTA (0.05%: 0.02%; w:v) for 5 min at 37 °C. Stop the enzymatic reaction by the addition of Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS) at 3:1 (v:v) ratio of added trypsin-EDTA.
NOTE: HeLa cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated FCS at 37 °C and 5% CO2 and were routinely passaged once they reached 80–90% confluency as described above. - Determine the cell concentration using a Neubauer chamber. Mix a 50 μL aliquot of the cell suspension with trypan blue at 1:1 (v:v) ratio, resuspend, and transfer 10 μL of the suspension into the chamber. Count only the trypan blue-negative cells inside of the four large squares (for details see Phelan et al.47). Derive the cell concentration from the counted cell number using the following formula:
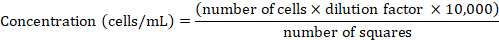
- Pellet the cells by centrifugation at 300 x g for 2 min. Resuspend with fresh RPMI 1640 in order to obtain 1 x 106 cells/mL.
- Remove the fibronectin from the chambered coverslip, wash the wells twice with DPBS, and seed 50,000 cells per well.
- Return the chambered coverslip with the cells to the incubator and grow them for 24 h at 37 °C and 5% CO2.
2. Expression of pEB3-tdTomato in HeLa Cells
- Prepare a 1.5 mL microcentrifuge tube. For each tube, dilute 2 μg of pEB3-tdTomato48 with transfection buffer (synthetic product in aqueous solution) to a final volume of 396 μL.
- Add 4 μL of transfection reagent (non-lipidic, containing polyethylenimine) to the first tube, and vortex the mixture immediately for exactly 10 s.
- Briefly spin down the tube with a microcentrifuge and incubate at room temperature (RT) for 10 min.
- Remove the HeLa cells from the incubator. Dropwise, add 100 μL of the transfection mixture to each well of a 4 well chambered coverslip, and return the cells to the incubator.
- After 4 h of incubation at 37 °C and 5% CO2, supplement the cells with fresh growth medium and incubate for at least 24 h at 37 °C and 5% CO2.
NOTE: It is necessary to optimize transfection conditions for each cell type. The expression levels need to be low enough to allow the identification of single MT growing ends. Alternatively, a cell line stably expressing EB3-tdTomato can be used in the experiments; this would reduce variability in expression levels of EB3-tdTomato between preparations and between cells from the same preparation49.
3. Synchronization and Live-cell Imaging of pEB3-tdTomato–expressing HeLa Cells
- Prepare a 2.5 μM solution of dimethylenastron (DME) in phenol-red free Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FCS and 2 mM L-glutamine or an alternative glutamine supply.
- Replace the growth medium in the chambered coverslip with 500 μL of the growth medium containing 2.5 μM DME and incubate the cells at 37 °C and 5% CO2.
- After 3.5 h of incubation with DME, transfer the cells to the microscope, mount the chambered coverslip into an environmental chamber with dark panels for imaging at 37 °C and 5% CO2, and further incubate until the total incubation time is 4 h.
NOTE: The maintenance of temperature at 37 °C without fluctuation is crucial for the experiment. - Perform the time-lapse imaging on an inverted microscope equipped with a 100x 1.49 N.A. oil immersion objective, a dual spinning disk confocal system, and a reliable autofocus system for continuous maintenance of the focal plane. Define the imaging parameters as follows.
NOTE: We use an Electron Multiplying Charge-Coupled Device camera (EM-CCD).- For EB3-tdTomato excitation, use a 561 nm laser line with 200 ms exposure time. Collect the emitted light through a quadruple bandpass (405, 488, 561, 640 nm) dichroic mirror and a 600/52 nm emission filter.
NOTE: Laser power can be adjusted for each imaged cell to prevent image saturation. In all time-lapse movies given here the laser power was set to 5.3 mW. - Find a cell in prophase and focus in the Z-plane corresponding to the center of the monopolar mitotic spindle. Acquire images every 0.5 s over a total of 1 min with no binning and no illumination between the exposures.
- For EB3-tdTomato excitation, use a 561 nm laser line with 200 ms exposure time. Collect the emitted light through a quadruple bandpass (405, 488, 561, 640 nm) dichroic mirror and a 600/52 nm emission filter.
4. Analysis of the MT Dynamics Using U-Track v2.2.0
- To analyze the MT dynamics a numerical computing environment software is required (e.g., MATLAB).
NOTE: Basic understanding of the software is sufficient for the analysis. Comprehensive help material and tutorials are available on the developer’s website (https://uk.mathworks.com/products/matlab/getting-started.html). - Download (https://github.com/DanuserLab/u-track) and install the open-source U-Track v2.2.0 software following the detailed instructions given in the "Readme_u-track.pdf" file50,51,52.
- Launch the numerical-analysis software and add U-Track v2.2.0 folder with subfolders into the software search path.
- From the command window call "movieSelectorGUI". This opens a dialogue window from which the raw files generated by the image acquisition software at the microscope can be imported (Supplementary Figure 1, Figure 2, Figure 3, Figure 4).
NOTE: The U-Track software is compatible with other image data formats. It uses Bio-Formats, which recognizes different life science data formats53. - The size of each image is read from the metadata automatically. Manually enter the numerical aperture of the objective (in this case 1.49) and the time interval (0.5 s) used for imaging (Supplementary Figure 1B). Additionally, information on the excitation wavelength, the fluorophore, and the exposure time can also be provided, but they are not critical for further analysis.
- Once all the images are loaded, save the entered time-lapse series as a movie list by selecting the "Save As Movie List". On the right side of the dialogue window select the "U-Track" option and press "Continue" (Supplementary Figure 1C).
NOTE: The values are optimized for HeLa cells. If switching to a different cell line, the values should be defined again. Alternatively, use the settings recommended by the software developers. The detailed explanation of each of the parameters and how they should be defined can be found in the technical report provided with the previous version of the software, plusTipTracker50. - From the pop-up window select "Microtubule Plus-Ends" and press "Ok" (Supplementary Figure 1C). The new dialogue window allows determining the parameters for the three steps of the analysis (Supplementary Figure 1D), which are detection, tracking, and track analysis.
- In step 1 choose "Settings" and from a drop-down menu select "Comet Detection" as a detection method (Supplementary Figure 2B).
- From the new dialogue window define the parameters for the difference of Gaussians filter and the watershed segmentation as follows (Supplementary Figure 2C): Mask process to be used for the detection = None; Low-pass Gaussian standard deviation = 1 pixel; High-pass Gaussian standard deviation = 3 pixels; Minimum threshold = 3 standard deviations; Threshold step size = 0.25 standard deviations. Select "Apply Settings to All Movies" and "Apply".
- In step 2, the parameters for linking, gap closing, merging and splitting, and Kalman filter functions are defined in three steps as highlighted in pink, green, and blue, accordingly (Supplementary Figure 3B). For these steps, select the "Microtubule Plus-end Dynamics" and from the "Setting" option define the values as indicated in Supplementary Figure 3C–E, respectively.
- For problems with dimensionality, choose "2" from the drop-down menu. Use Maximum Gap to Close = 5 frames; Minimum Length of Track Segments from First Step = 3 frames. As before, select "Apply Settings to All Movies" and click on "Apply".
- In step 3 of the analysis, the detected MT tracks are classified (Supplementary Figure 4). As a track analysis method, choose "Microtubule Dynamics Classification" and define the parameters through the "Setting" button as indicated in Supplementary Figure 4B,C. After that, choose the "Apply Settings to All Movies" box and click on "Apply".
- Once all the parameters are defined, from the "Control Panel–U-Track" window (Supplementary Figure 1D) select the "Apply Check/Uncheck to All Movies" and "Run All Movies" boxes and press "Run". This will initiate the MT analysis of the time-lapse series.
- Once the movie processing is completed, a message "Your movie(s) have been processed successfully" is displayed. Press "Ok", then "Save".
- Now it is safe to quit the numerical-analysis software. The results from the movie processing are stored in subfolder structures as m-files in the folder where the raw files are stored.
5. Statistical Analysis of the MT Dynamics
- Import the m-files into a preferred statistic analysis program.
NOTE: In our case, we first import the files in a standard spreadsheet to make them readable. The m-files contain statistical information (median, mean, and standard deviation) on different parameters (e.g., growth speed, MT dynamicity). The detailed list of the parameters is given in the technical report provided with the previous version of the software, plusTipTracker50,52. The generated m-files can also be imported into other data processing software. - Choose the "growth speed mean" parameter and import it into a table for statistics and display. Enter the information on other parameters, (e.g., "dynamicity") either in a new table or in a new column of the same grouped table and plot.
Access restricted. Please log in or start a trial to view this content.
Results
Following the given protocol outlined in Figure 1A, the pEB3-tdTomato plasmid was transiently expressed in asynchronously growing HeLa cells. The cells were synchronized 48 h after the transfection at prometaphase through DME treatment (Figure 1B). This step ensured that the measurement of MT dynamics was always done at the same phase of the cell cycle. The time-lapse movies were further processed and analyzed with U-Track v2.2.0 as described in its supplementar...
Access restricted. Please log in or start a trial to view this content.
Discussion
Here, we describe a modification of a method first established by Ertych et al.44. Along with several other modifications, we combine this technique of MT dynamics analysis with dual spinning disk confocal imaging. The use of the dual spinning disk improves the resolution of growing MTs while reducing phototoxicity36. We further reduce the photobleaching and laser light-induced damage of the cells by switching to a longer wavelength fluorescent reporter. The tdTomato fluore...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank the members of the Light Microscopy Facility, Max-Planck Institute of Experimental Medicine, for their expert advice and support.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Dimethylenastron | Merck | 324622 | |
| DMEM w/o phenol red | Gibco | 31053-28 | |
| DPBS | Gibco | 14190-094 | |
| Fetal bovine serum | Biochrom | S0415 | |
| Fibronectin Bovine Plasma | Merck | F4759 | Sterile powder |
| GlutaMAX | Gibco | 35050-038 | Stable glutamine substitutive |
| jetPRIME | Polyplus | 114-15 | |
| EB3-TdTomato | Addgene | plasmid #50708 | |
| RPMI 1640 | Gibco | 61870-010 | |
| Trypan Blue | Merck | T8154-20ML | |
| Trypsin/EDTA solution | Biochrom | L2143 | 0.05%/0.02 % w/o calcium and magnesium |
| µ-slide | Ibidi | 80426 | 4-well slide with #1.5 coverslip |
| Eclipse Ti Inverted microscope | Nikon | NA | |
| Objective | Nikon | MRD01991 | CFI Apo TIRF 100xC Oil |
| ACAL Laser Excahnger | Nikon | Laser box. 405, 458, 488, 514, 561 and 647 nm | |
| Spinning disk module | Andor | CSU-W | |
| Camera | Andor | iXon Ultra 888 | |
| Environmental Chamber | Okolab | Dark chamber equipped with CO2 supply, temperature control and humidifier | |
| HeLa Cells | DSMZ | ACC-57 | |
| NIS Elements v4 | Nikon | Spinning disk microscope. Acquisition Software | |
| MATLAB | Mathworks | Computing environment | |
| Prism 8 | GraphPad | Statistical analysis and display software |
References
- Erickson, H. P. Evolution of the cytoskeleton. Bioessays. 29 (7), 668-677 (2007).
- Pollard, T. D., Goldman, R. D. Overview of the Cytoskeleton from an Evolutionary Perspective. Cold Spring Harbor Perspectives in Biology. 10 (7), (2018).
- Wade, R. H. On and around microtubules: an overview. Molecular Biotechnology. 43 (2), 177-191 (2009).
- Forth, S., Kapoor, T. M. The mechanics of microtubule networks in cell division. Journal of Cell Biology. 216 (6), 1525-1531 (2017).
- Franker, M. A., Hoogenraad, C. C. Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. Journal of Cell Science. 126, Pt 11 2319-2329 (2013).
- Lindemann, C. B., Lesich, K. A. Flagellar and ciliary beating: the proven and the possible. Journal of Cell Science. 123, Pt 4 519-528 (2010).
- Wheway, G., Nazlamova, L., Hancock, J. T. Signaling through the Primary Cilium. Frontiers in Cell and Developmental Biology. 6, 8(2018).
- Falk, N., Losl, M., Schroder, N., Giessl, A. Specialized Cilia in Mammalian Sensory Systems. Cells. 4 (3), 500-519 (2015).
- Spoon, C., Grant, W. Biomechanical measurement of kinocilium. Methods in Enzymology. 525, 21-43 (2013).
- Zenker, J., et al. A microtubule-organizing center directing intracellular transport in the early mouse embryo. Science. 357 (6354), 925-928 (2017).
- Goldstein, B. Embryonic polarity: a role for microtubules. Current Biology. 10 (22), 820-822 (2000).
- Uchida, S., Shumyatsky, G. P. Deceivingly dynamic: Learning-dependent changes in stathmin and microtubules. Neurobiology of Learning and Memory. 124, 52-61 (2015).
- Fife, C. M., McCarroll, J. A., Kavallaris, M. Movers and shakers: cell cytoskeleton in cancer metastasis. British Journal of Pharmacology. 171 (24), 5507-5523 (2014).
- Bouchet, B. P., Akhmanova, A. Microtubules in 3D cell motility. Journal of Cell Science. 130 (1), 39-50 (2017).
- Dent, E. W. Of microtubules and memory: implications for microtubule dynamics in dendrites and spines. Molecular Biology of the Cell. 28 (1), 1-8 (2017).
- Craddock, T. J., Tuszynski, J. A., Hameroff, S. Cytoskeletal signaling: is memory encoded in microtubule lattices by CaMKII phosphorylation. PLOS Computational Biology. 8 (3), (2012).
- Smythies, J. Off the beaten track: the molecular structure of long-term memory: three novel hypotheses-electrical, chemical and anatomical (allosteric). Frontiers in Integrative Neuroscience. 9, 4(2015).
- Kaganovsky, K., Wang, C. Y. How Do Microtubule Dynamics Relate to the Hallmarks of Learning and Memory. Journal of Neuroscience. 36 (22), 5911-5913 (2016).
- Mitchison, T., Kirschner, M. Dynamic instability of microtubule growth. Nature. 312 (5991), 237-242 (1984).
- Dubey, J., Ratnakaran, N., Koushika, S. P. Neurodegeneration and microtubule dynamics: death by a thousand cuts. Frontiers in Cellular Neuroscience. 9, 343(2015).
- Parker, A. L., Kavallaris, M., McCarroll, J. A. Microtubules and their role in cellular stress in cancer. Frontiers in Oncology. 4, 153(2014).
- Honore, S., Pasquier, E., Braguer, D. Understanding microtubule dynamics for improved cancer therapy. Cell and Molecular Life Sciences. 62 (24), 3039-3056 (2005).
- Straube, A. Methods in Molecular Biology. , Humana Press. Totowa, NJ. (2011).
- Budde, P. P., Desai, A., Heald, R. Analysis of microtubule polymerization in vitro and during the cell cycle in Xenopus egg extracts. Methods. 38 (1), 29-34 (2006).
- Gierke, S., Kumar, P., Wittmann, T. Analysis of microtubule polymerization dynamics in live cells. Methods in Cell Biology. 97, 15-33 (2010).
- Matov, A., et al. Analysis of microtubule dynamic instability using a plus-end growth marker. Nature Methods. 7 (9), 761-768 (2010).
- Bailey, M., Conway, L., Gramlich, M. W., Hawkins, T. L., Ross, J. L. Modern methods to interrogate microtubule dynamics. Integrative Biology (Camb). 5 (11), 1324-1333 (2013).
- Galjart, N. Plus-end-tracking proteins and their interactions at microtubule ends. Current Biology. 20 (12), 528-537 (2010).
- Stepanova, T., et al. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). Journal of Neuroscience. 23 (7), 2655-2664 (2003).
- Zwetsloot, A. J., Tut, G., Straube, A. Measuring microtubule dynamics. Essays in Biochemistry. 62 (6), 725-735 (2018).
- Bayguinov, P. O., et al. Modern Laser Scanning Confocal Microscopy. Current Protocols in Cytometry. 85 (1), 39(2018).
- Nakano, A. Spinning-disk confocal microscopy -- a cutting-edge tool for imaging of membrane traffic. Cell Structure and Function. 27 (5), 349-355 (2002).
- Elektrisches teleskop. Germany patent. , (1884).
- Yin, S., Lu, G., Zhang, J., Yu, F. T., Mait, J. N. Kinoform-based Nipkow disk for a confocal microscope. Applied Optics. 34 (25), 5695-5698 (1995).
- Nipkow disk for confocal optical scanner. European patent application. , EP92114750A (1992).
- Oreopoulos, J., Berman, R., Browne, M. Spinning-disk confocal microscopy: present technology and future trends. Methods in Cell Biology. 123, 153-175 (2014).
- Rusan, N. M., Fagerstrom, C. J., Yvon, A. M., Wadsworth, P. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Molecular Biology of the Cell. 12 (4), 971-980 (2001).
- Rusan, N. M., Fagerstrom, C. J., Yvon, A. -M. C., Wadsworth, P. Cell Cycle-Dependent Changes in Microtubule Dynamics in Living Cells Expressing Green Fluorescent Protein-α Tubulin. Molecular Biology of the Cell. 12 (4), 971-980 (2001).
- Liu, D., Davydenko, O., Lampson, M. A. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. Journal of Cell Biology. 198 (4), 491-499 (2012).
- Maiato, H., Sunkel, C. E. Kinetochore-microtubule interactions during cell division. Chromosome Research. 12 (6), 585-597 (2004).
- Muller, C., et al. Inhibitors of kinesin Eg5: antiproliferative activity of monastrol analogues against human glioblastoma cells. Cancer Chemotherrapy and Pharmacology. 59 (2), 157-164 (2007).
- Mayer, T. U., et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 286 (5441), 971-974 (1999).
- Kapoor, T. M., Mayer, T. U., Coughlin, M. L., Mitchison, T. J. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin Eg5. The Journal of Cell Biology. 150 (5), 975-988 (2000).
- Ertych, N., et al. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nature Cell Biology. 16 (8), 779-791 (2014).
- Brito, D. A., Yang, Z., Rieder, C. L. Microtubules do not promote mitotic slippage when the spindle assembly checkpoint cannot be satisfied. The Journal of Cell Biology. 182 (4), 623-629 (2008).
- Shaner, N. C., Patterson, G. H., Davidson, M. W. Advances in fluorescent protein technology. Journal of Cell Science. 120 (24), 4247-4260 (2007).
- Phelan, M. C., Lawler, G. Cell Counting. Current Protocols in Cytometry. 00 (1), 3(1997).
- Merriam, E. B., et al. Synaptic regulation of microtubule dynamics in dendritic spines by calcium, F-actin, and drebrin. Journal of Neuroscience. 33 (42), 16471-16482 (2013).
- Samora, C. P., et al. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nature Cell Biology. 13 (9), 1040-1050 (2011).
- Applegate, K. T., et al. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. Journal of Structural Biology. 176 (2), 168-184 (2011).
- Jaqaman, K., et al. Robust single-particle tracking in live-cell time-lapse sequences. Nature Methods. 5 (8), 695-702 (2008).
- Stout, A., D'Amico, S., Enzenbacher, T., Ebbert, P., Lowery, L. A. Using plusTipTracker Software to Measure Microtubule Dynamics in Xenopus laevis Growth Cones. Journal of Visualized Experiments. , e52138(2014).
- Linkert, M., et al. Metadata matters: access to image data in the real world. The Journal of Cell Biology. 189 (5), 777-782 (2010).
- Brouhard, G. J. Dynamic instability 30 years later: complexities in microtubule growth and catastrophe. Molecular Biology of the Cell. 26 (7), 1207-1210 (2015).
- Burbank, K. S., Mitchison, T. J. Microtubule dynamic instability. Current Biology : CB. 16 (14), 516-517 (2006).
- Caplow, M., Shanks, J., Ruhlen, R. L. Temperature-jump studies of microtubule dynamic instability. Journal of Biological Chemistry. 263 (21), 10344-10352 (1988).
- Prasad, V., Jordan, M. A., Luduena, R. F. Temperature sensitivity of vinblastine-induced tubulin polymerization in the presence of microtubule-associated proteins. Journal of Protein Chemistry. 11 (5), 509-515 (1992).
- Wasteneys, G. O. Microtubules Show their Sensitive Nature. Plant and Cell Physiology. 44 (7), 653-654 (2003).
- Turi, A., Lu, R. C., Lin, P. -S. Effect of heat on the microtubule disassembly and its relationship to body temperatures. Biochemical and Biophysical Research Communications. 100 (2), 584-590 (1981).
- Safinya, C. R., et al. The effect of multivalent cations and Tau on paclitaxel-stabilized microtubule assembly, disassembly, and structure. Advances in Colloid and Interface Science. 232, 9-16 (2016).
- Sandoval, I. V., Weber, K. Calcium-Induced Inactivation of Microtubule Formation in Brain Extracts. European Journal of Biochemistry. 92 (2), 463-470 (1978).
- Vater, W., Böhm, K. J., Unger, E. Tubulin assembly in the presence of calcium ions and taxol: Microtubule bundling and formation of macrotubule-ring complexes. Cell Motility. 36 (1), 76-83 (1997).
- Yamashita, N., et al. Three-dimensional tracking of plus-tips by lattice light-sheet microscopy permits the quantification of microtubule growth trajectories within the mitotic apparatus. Journal of Biomedical Optics. 20 (10), 1-18 (2015).
- Pamula, M. C., et al. High-resolution imaging reveals how the spindle midzone impacts chromosome movement. Journal of Cell Biology. 218 (8), 2529-2544 (2019).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved