A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Swin-PSAxialNet: An Efficient Multi-Organ Segmentation Technique
In This Article
Summary
The present protocol describes an efficient multi-organ segmentation method called Swin-PSAxialNet, which has achieved excellent accuracy compared to previous segmentation methods. The key steps of this procedure include dataset collection, environment configuration, data preprocessing, model training and comparison, and ablation experiments.
Abstract
Abdominal multi-organ segmentation is one of the most important topics in the field of medical image analysis, and it plays an important role in supporting clinical workflows such as disease diagnosis and treatment planning. In this study, an efficient multi-organ segmentation method called Swin-PSAxialNet based on the nnU-Net architecture is proposed. It was designed specifically for the precise segmentation of 11 abdominal organs in CT images. The proposed network has made the following improvements compared to nnU-Net. Firstly, Space-to-depth (SPD) modules and parameter-shared axial attention (PSAA) feature extraction blocks were introduced, enhancing the capability of 3D image feature extraction. Secondly, a multi-scale image fusion approach was employed to capture detailed information and spatial features, improving the capability of extracting subtle features and edge features. Lastly, a parameter-sharing method was introduced to reduce the model's computational cost and training speed. The proposed network achieves an average Dice coefficient of 0.93342 for the segmentation task involving 11 organs. Experimental results indicate the notable superiority of Swin-PSAxialNet over previous mainstream segmentation methods. The method shows excellent accuracy and low computational costs in segmenting major abdominal organs.
Introduction
Contemporary clinical intervention, including the diagnosis of diseases, the formulation of treatment plans, and the tracking of treatment outcomes, relies on the accurate segmentation of medical images1. However, the complex structural relationships among abdominal organs2make it a challenging task to achieve accurate segmentation of multiple abdominal organs3. Over the past few decades, the flourishing developments in medical imaging and computer vision have presented both new opportunities and challenges in the field of abdominal multi-organ segmentation. Advanced Magnetic Resonance Imaging (MRI)4 and Computed Tomography (CT) technology5 enable us to acquire high-resolution abdominal images. The precise segmentation of multiple organs from CT images holds significant clinical value for the assessment and treatment of vital organs such as the liver, kidneys, spleen, pancreas, etc.6,7,8,9,10 However, manual annotation of these anatomical structures, especially those requiring intervention from radiologists or radiation oncologists, is both time-consuming and susceptible to subjective influences11. Therefore, there is an urgent need to develop automated and accurate methods for abdominal multi-organ segmentation.
Previous research on image segmentation predominantly relied on Convolutional Neural Networks (CNNs), which improve segmentation efficiency by stacking layers and introducing ResNet12. In 2020, the Google research team introduced the Vision Transformer (VIT) model13, marking a pioneering instance of incorporating Transformer architecture into the traditional visual domain for a range of visual tasks14. While convolutional operations can only contemplate local feature information, the attention mechanism in Transformers enables the comprehensive consideration of global feature information.
Considering the superiority of Transformer-based architectures over traditional convolutional networks15, numerous research teams have undertaken extensive exploration in optimizing the synergy between the strengths of Transformers and convolutional networks16,17,18,19. Chen et al. introduced the TransUNet for medical image segmentation tasks16, which leverages Transformers to extract global features from images. Due to the high cost of network training and the failure to utilize the concept of feature extraction hierarchy, the advantages of Transformer have not been fully realized.
To address these issues, many researchers have started experimenting with incorporating Transformers as the backbone for training segmentation networks. Liu et al.17 introduced the Swin Transformer, which employed a hierarchical construction method for layered feature extraction. The concept of Windows Multi-Head Self-Attention (W-MSA) was proposed, significantly reducing computational cost, particularly in the presence of larger shallow-level feature maps. While this approach reduced computational requirements, it also isolated information transmission between different windows. To address this problem, the authors further introduced the concept of Shifted Windows Multi-Head Self-Attention (SW-MSA), enabling information propagation among adjacent windows. Building upon this methodology, Cao et al. formulated the Swin-UNet18, replacing the 2D convolutions in U-Net with Swin modules and incorporating W-MSA and SW-MSA into the encoding and decoding processes, achieving commendable segmentation outcomes.
Conversely, Zhou et al. highlighted that the advantage of conv operation could not be ignored when processing high-resolution images19. Their proposed nnFormer employs a self-attention computation method based on local three-dimensional image blocks, constituting a Transformer model characterized by a cross-shaped structure. The utilization of attention based on local three-dimensional blocks significantly reduced the training load on the network.
Given the problems with the above study, an efficient hybrid hierarchical structure for 3D medical image segmentation, termed Swin-PSAxialNet, is proposed. This method incorporates a downsampling block, Space-to-depth (SPD)20 block, capable of extracting global information21. Additionally, it adds a parameter shared axial attention (PSAA) module, which reduces the learning parameter count from quadratic to linear and will have a good effect on the accuracy of network training and the complexity of training models22.
Swin-PSAxialNet network
The overall architecture of the network adopts the U-shaped structure of nnU-Net23, consisting of encoder and decoder structures. These structures engage in local feature extraction and the concatenation of features from large and small-scale images, as illustrated in Figure 1.
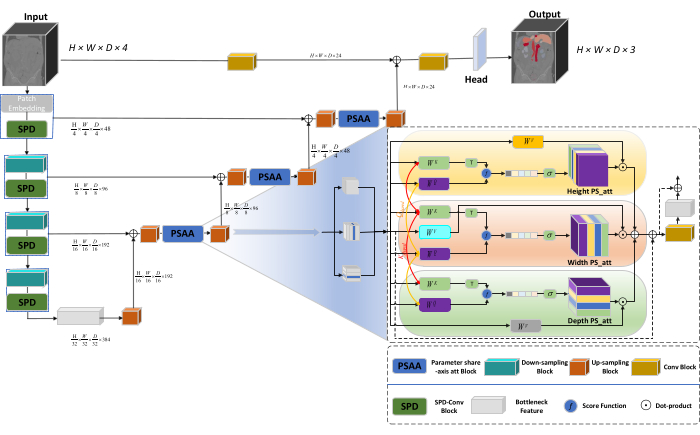
Figure 1: Swin-PSAxialNet schematic diagram of network architecture. Please click here to view a larger version of this figure.
In the encoder structure, the traditional Conv block is combined with the SPD block20 to form a downsampling volume. The first layer of the encoder incorporates Patch Embedding, a module that partitions the 3D data into 3D patches 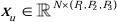 , (P1, P2, P3) represents non-overlapping patches in this context,
, (P1, P2, P3) represents non-overlapping patches in this context, 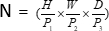 signifies the sequence length of 3D patches. Following the embedding layer, the next step involves a non-overlapping convolutional downsampling unit comprising both a convolutional block and an SPD block. In this setup, the convolutional block has a stride set to 1, and the SPD block is employed for image scaling, leading to a fourfold reduction in resolution and a twofold increase in channels.
signifies the sequence length of 3D patches. Following the embedding layer, the next step involves a non-overlapping convolutional downsampling unit comprising both a convolutional block and an SPD block. In this setup, the convolutional block has a stride set to 1, and the SPD block is employed for image scaling, leading to a fourfold reduction in resolution and a twofold increase in channels.
In the decoder structure, each upsample block after the Bottleneck Feature layer consists of a combination of an upsampling block and a PSAA block. The resolution of the feature map is increased twofold, and the channel count is halved between each pair of decoder stages. To restore spatial information and enhance feature representation, feature fusion between large and small-scale images is performed between the upsampling blocks. Ultimately, the upsampling results are fed into the Head layer to restore the original image size, with an output size of (H × W × D × C, C = 3).
SPD block architecture
In traditional methods, the downsampling section employs a single stride with a step size of 2. This involves convolutional pooling at local positions in the image, limiting the receptive field, and confining the model to the extraction of features from small image patches. This method utilizes the SPD block, which finely divides the original image into three dimensions. The original 3D image is evenly segmented along the x, y, and z axes, resulting in four sub-volume bodies. (Figure 2) Subsequently, the four volumes are concatenated through "cat" operation, and the resulting image undergoes a 1 × 1 × 1 convolution to obtain the down-sampled image20.
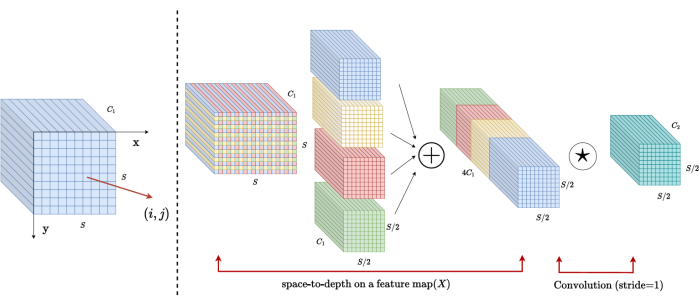
Figure 2: SPD block diagram. Please click here to view a larger version of this figure.
PSAA block architecture
In contrast to traditional CNN networks, the proposed PSAA block is more effective in conducting global information focus and more efficient in network learning and training. This enables the capture of richer images and spatial features. The PSAA block includes axial attention learning based on params sharing in three dimensions: Height, Width, and Depth. In comparison to the conventional attention mechanism that performs attention learning for each pixel in the image, this method independently conducts attention learning for each of the three dimensions, reducing the complexity of self-attention from quadratic to linear. Moreover, a learnable keys-queries parameter sharing mechanism is employed, enabling the network to perform attention mechanism operations in parallel across the three dimensions, resulting in faster, superior, and more effective feature representation.
Access restricted. Please log in or start a trial to view this content.
Protocol
The present protocol was approved by the Ethics Committee of Nantong University. It involves the intelligent assessment and research of acquired non-invasive or minimally invasive multimodal data, including human medical images, limb movements, and vascular imaging, utilizing artificial intelligence technology. Figure 3 depicts the overall flowchart of multi-organ segmentation. All the necessary weblinks are provided in the Table of Materials.
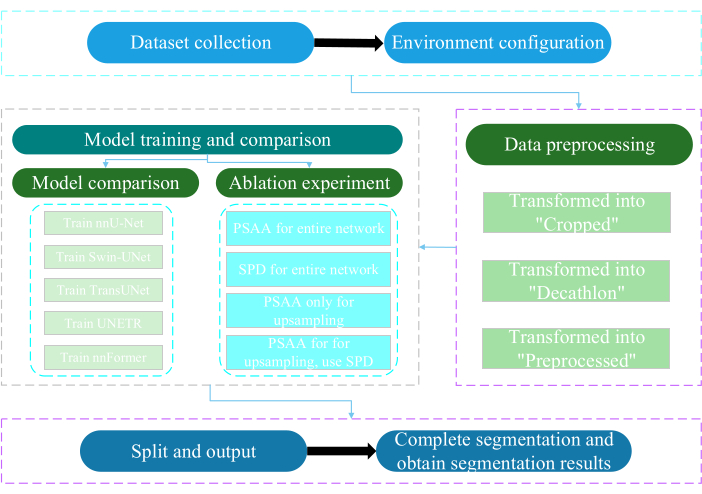
Figure 3: Overall flowchart of the multi-organ segmentation. Please click here to view a larger version of this figure.
1. Dataset collection
- Download the AMOS2022 dataset24 (see Table of Materials).
- Select 160 images as the training dataset and 40 images as the testing dataset.
- Terminally execute mkdir ~/PaddleSeg/contrib/MedicalSeg/data/raw_data.
- Open the path of raw_data.
- Place the dataset in the raw_data directory, and ensure correct correspondence with the file names in the "imagesTr" and "labelsTr" directories.
- Create dataset.json file.
- Add dataset information such as numTraining, numTest, and labels name into the "dataset.json" file.
NOTE: The dataset for this study consists of 200 high-quality CT datasets selected from the AMOS2022 official 500 CT dataset for training and testing purposes. The dataset encompasses 11 organs, including the spleen, right kidney, left kidney, gallbladder, esophagus, liver, stomach, aorta, inferior vena cava, pancreas, and bladder.
2. Environment configuration
- Install the Paddlepaddle environment (see Table of Materials) and the corresponding CUDA version25.
NOTE: Utilizing Paddlepaddle version 2.5.0 and CUDA version 11.7 is recommended. - Git clone the PaddleSeg repository25.
- Terminally execute cd ~/PaddleSeg/contrib/MedicalSeg/.
- Execute pip install -r requirements.txt.
- Execute pip install medpy.
3. Data preprocessing
- Modify the "yml" file, including parameters such as data_root, batch_size, model, train_dataset, val_dataset, optimizer, lr_scheduler, and loss.
- Specify the required parameters for execution, including "-config, --log_iters, --precision, --nnunet, --save_dir, --save_interval, etc.
NOTE: The original "raw_data" is transformed into three types of data files: "Cropped," "Decathlon," and "Preprocessed," catering to different training and evaluation requirements to enhance model performance and training effectiveness.
4. Model training and comparison
NOTE: As a widely used baseline in the field of image segmentation, nnU-Net23 serves as a baseline model in the study. The specific model comparison process is as follows.
- Train nnU-Net
- Configure with a batch size of 2 and 80,000 iterations, log every 20 iterations, and utilize fp16 precision.
- Set the save_dir as the "output" folder within the working directory, with a save interval set at 1,000 iterations.
- Add parameter --use_vdl for VisualDL parameter visualization.
- Run train.py.
- Create four yml files, specify folds 1-4, and train the model with five-fold cross-validation.
- Train Swin-UNet
- Maintain consistency with the previously established experimental conditions.
- Remove parameter --nnunet and use --swinunet.
- Run the train.py file.
- Train folds 1-4 individually and then subject to ensemble processing.
- Train TransUNet
- Follow the procedures outlined in step 4.1.
- Open yml file.
- Modify type: nnunet to type: transunet.
- Modify parameter --nnunet to --transunet.
- Run the train.py file.
- Train folds 1-4 individually and process ensemble processing.
- Train UNETR
- Maintain consistent experimental variables.
- Repeat the procedures outlined previously like Swin-UNet and TransUNet (step 4.2 and step 4.3).
- Run train.py.
- Train folds 1-4 individually and process ensemble processing.
- Train nnFormer
- Modify the parameters like Swin-UNet and TransUNet.
- Run single fold train.py.
- Run ensemble processing.
5. Ablation experiment
- PSAA block for the entire network
- Keep variables consistent with step 4.1.
- Set volume_shape = (64,192,192) and Axial_attention = True.
- Run single fold train.py.
- Process ensemble processing across five folds.
- SPD block for the entire network
- Keep variables consistent with step 4.1.
- Set SPD_enable = True and Axial_attention = False.
- Run single fold train.py.
- Process ensemble processing across five folds.
- PSAA block for upsampling
NOTE: Do not use SPD block for downsampling.- Use the PSAA block for upsampling.
- Keep variables consistent with step 4.1.
- Set Axial_attention = False in the downsampling part and SPD_enable = False.
- Run single fold train.py.
- Process five folds ensemble processing.
- PSAA block for upsampling
NOTE: Use SPD block for downsampling.- Use the SPD block for downsampling and the PSAA block for upsampling.
- Keep variables consistent with step 4.1.
- Set Axial_attention = False in the downsampling part and SPD_enable = True.
- Run single fold train.py.
- Process five folds ensemble processing.
- Set the number of iterations to 2,00,000 because of the superior results.
Access restricted. Please log in or start a trial to view this content.
Results
This protocol employs two metrics to evaluate the model: Dice Similarity Score (DSC) and 95% Hausdorff Distance (HD95). DSC measures the overlap between voxel segmentation predictions and ground truth, while 95% HD assesses the overlap between voxel segmentation prediction boundaries and ground truth, filtering out 5% of outliers. The definition of DSC26 is as follows:
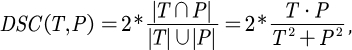 ...
...
Access restricted. Please log in or start a trial to view this content.
Discussion
The segmentation of abdominal organs is a complicated work. Compared to other internal structures of the human body, such as the brain or heart, segmenting abdominal organs seems more challenging because of the low contrast and large shape changes in CT images27,28. Swin-PSAxialNet is proposed here to solve this difficult problem.
In the data collection step, this study downloaded 200 images from the AMOS2022 official website
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors declare no conflicts of interest.
Acknowledgements
This study was supported by the '333' Engineering Project of Jiangsu Province ([2022]21-003), the Wuxi Health Commission General Program (M202205), and the Wuxi Science and Technology Development Fund (Y20212002-1), whose contributions have been invaluable to the success of this work." The authors thank all the research assistants and study participants for their support.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| AMOS2022 dataset | None | None | Datasets for network training and testing. The weblink is: https://pan.baidu.com/s/1x2ZW5FiZtVap0er55Wk4VQ?pwd=xhpb |
| ASUS mainframe | ASUS | https://www.asusparts.eu/en/asus-13020-01910200 | |
| CUDA version 11.7 | NVIDIA | https://developer.nvidia.com/cuda-11-7-0-download-archive | |
| NVIDIA GeForce RTX 3090 | NVIDIA | https://www.nvidia.com/en-in/geforce/graphics-cards/30-series/rtx-3090-3090ti/ | |
| Paddlepaddle environment | Baidu | None | Environmental preparation for network training. The weblink is: https://www.paddlepaddle.org.cn/ |
| PaddleSeg | Baidu | None | The baseline we use: https://github.com/PaddlePaddle/PaddleSeg |
References
- Liu, X., Song, L., Liu, S., Zhang, Y. A review of deep-learning-based medical image segmentation methods. Sustain. 13 (3), 1224(2021).
- Popilock, R., Sandrasagaren, K., Harris, L., Kaser, K. A. CT artifact recognition for the nuclear technologist. J Nucl Med Technol. 36 (2), 79-81 (2008).
- Rehman, A., Khan, F. G. A deep learning based review on abdominal images. Multimed Tools Appl. 80, 30321-30352 (2021).
- Schenck, J. F. The role of magnetic susceptibility in magnetic resonance imaging: Mri magnetic compatibility of the first and second kinds. Med Phys. 23 (6), 815-850 (1996).
- Palmisano, A., et al. Myocardial late contrast enhancement ct in troponin-positive acute chest pain syndrome. Radiol. 302 (3), 545-553 (2022).
- Chen, E. -L., Chung, P. -C., Chen, C. -L., Tsai, H. -M., Chang, C. -I. An automatic diagnostic system for CT liver image classification. IEEE Trans Biomed Eng. 45 (6), 783-794 (1998).
- Sagel, S. S., Stanley, R. J., Levitt, R. G., Geisse, G. J. R. Computed tomography of the kidney. Radiol. 124 (2), 359-370 (1977).
- Freeman, J. L., Jafri, S., Roberts, J. L., Mezwa, D. G., Shirkhoda, A. CT of congenital and acquired abnormalities of the spleen. Radiographics. 13 (3), 597-610 (1993).
- Almeida, R. R., et al. Advances in pancreatic CT imaging. AJR Am J Roentgenol. 211 (1), 52-66 (2018).
- Eisen, G. M., et al. Guidelines for credentialing and granting privileges for endoscopic ultrasound. Gastrointest Endosc. 54 (6), 811-814 (2001).
- Sykes, J. Reflections on the current status of commercial automated segmentation systems in clinical practice. J Med Radiat Sci. 61 (3), 131-134 (2014).
- He, K., Zhang, X., Ren, S., Sun, J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). , Vegas, NV, USA. 770-778 (2016).
- Dosovitskiy, A., et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv. arXiv. , 11929(2020).
- Ramachandran, P., et al. Stand-alone self-attention in vision models. Adv Neural Inf Process Syst. 32, NIPS2019 (2019).
- Vaswani, A., et al. Attention is all you need. Adv Neural Inf Process Syst. 30, NIPS2019 (2017).
- Chen, J., et al. TransUNET: Transformers make strong encoders for medical image segmentation. arXiv. , arXiv:2102 04396(2021).
- Liu, Z., et al. Swin transformer: Hierarchical vision transformer using shifted windows. Proc IEEE Int Conf Comput Vis. , 10012-10022 (2021).
- Cao, H., et al. Swin-UNET: Unet-like pure transformer for medical image segmentation. Comput Vis ECCV. , 205-218 (2022).
- Zhou, H. -Y., et al. nnformer: Interleaved transformer for volumetric segmentation. arXiv. arXiv. , arXiv:2109 03201(2021).
- Sunkara, R., Luo, T. No more strided convolutions or pooling: A new CNN building block for low-resolution images and small objects. Mach Learn Knowl Discov Databases. , 443-459 (2023).
- Çiçek, Ö, Abdulkadir, A., Lienkamp, S. S., Brox, T., Ronneberger, O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. Med Comput Vis Bayesian Graph Models Biomed Imaging. 2016, 424-432 (2016).
- Kim, H., et al. Abdominal multi-organ auto-segmentation using 3D-patch-based deep convolutional neural network. Scientific reports. 10 (1), 6204(2020).
- Isensee, F., et al. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv. , arXiv:1809 10486(2018).
- Ji, Y., et al. A large-scale abdominal multi-organ benchmark for versatile medical image segmentation. Adv Neural Inf Process Syst. 35, 36722-36732 (2022).
- Liu, Y., et al. Paddleseg: A high-efficient development toolkit for image segmentation. arXiv. , arXiv:2101 06175(2021).
- Hatamizadeh, A., et al. UNETR: Transformers for 3D medical image segmentation. Proc IEEE Int Conf Comput Vis. , https://api.semanticscholar.org/CorpusID:232290634 574-584 (2022).
- Zhou, H., Zeng, D., Bian, Z., Ma, J. A semi-supervised network-based tissue-aware contrast enhancement method for ct images. Nan Fang Yi Ke Da Xue Xue Bao. 43 (6), 985-993 (2023).
- Ma, J., et al. Abdomenct-1k: Is abdominal organ segmentation a solved problem. IEEE Trans Pattern Anal Mach Intell. 44 (10), 6695-6714 (2021).
- Galván, E., Mooney, P. Neuroevolution in deep neural networks: Current trends and future challenges. IEEE Trans Artif Intell. 2 (6), 476-493 (2021).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved